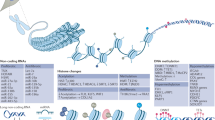
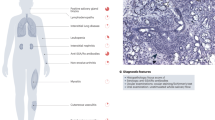
Abstract
Systemic sclerosis (SSc) is a severe autoimmune disease that is characterized by vascular abnormalities, immunological alterations and fibrosis of the skin and internal organs. The results of genetic studies in patients with SSc have revealed statistically significant genetic associations with disease manifestations and progression. Nevertheless, genetic susceptibility to SSc is moderate, and the functional consequences of genetic associations remain only partially characterized. A current hypothesis is that, in genetically susceptible individuals, epigenetic modifications constitute the driving force for disease initiation. As epigenetic alterations can occur years before fibrosis appears, these changes could represent a potential link between inflammation and tissue fibrosis. Epigenetics is a fast-growing discipline, and a considerable number of important epigenetic studies in SSc have been published in the past few years that span histone post-translational modifications, DNA methylation, microRNAs and long non-coding RNAs. This Review describes the latest insights into genetic and epigenetic contributions to the pathogenesis of SSc and aims to provide an improved understanding of the molecular pathways that link inflammation and fibrosis. This knowledge will be of paramount importance for the development of medicines that are effective in treating or even reversing tissue fibrosis.
Key points
-
Systemic sclerosis (SSc) is a complex fibrotic, autoimmune disease, the manifestations of which are only partially explained by genetic predisposition.
-
The concordance rate for SSc in monozygotic twins is low, indicating that genetic predisposition is insufficient to explain disease development and suggesting a role for environmental factors and epigenetic influences.
-
Epigenetic factors associated with SSc include changes in DNA methylation, histone modifications and the expression of microRNAs and long non-coding RNAs, which together drive aberrant immune activation and fibrosis.
-
Integration of the knowledge derived from genomic and epigenomic studies in SSc is needed to improve the characterization of the disease.
-
Therapies that target epigenetic pathways are emerging as promising therapeutic tools in experimental models of fibrosis, raising hope for future applications in SSc.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gabrielli, A., Avvedimento, E. V. & Krieg, T. Scleroderma. N. Engl. J. Med. 360, 1989–2003 (2009).
[No authors listed]. Preliminary criteria for the classification of systemic sclerosis (scleroderma). Subcommittee for scleroderma criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Arthritis Rheum. 23, 581–590 (1980).
Barnes, J. & Mayes, M. D. Epidemiology of systemic sclerosis: incidence, prevalence, survival, risk factors, malignancy, and environmental triggers. Curr. Opin. Rheumatol. 24, 165–170 (2012).
LeRoy, E. C. & Medsger, T. A. Jr. Criteria for the classification of early systemic sclerosis. J. Rheumatol. 28, 1573–1576 (2001).
van den Hoogen, F. et al. 2013 classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann. Rheum. Dis. 72, 1747–1755 (2013).
Pope, J. E. & Johnson, S. R. New classification criteria for systemic sclerosis (scleroderma). Rheum. Dis. Clin. North Am. 41, 383–398 (2015).
Chifflot, H., Fautrel, B., Sordet, C., Chatelus, E. & Sibilia, J. Incidence and prevalence of systemic sclerosis: a systematic literature review. Semin. Arthritis Rheum. 37, 223–235 (2008).
Roberts-Thomson, P. J. et al. Scleroderma in South Australia: epidemiological observations of possible pathogenic significance. Intern. Med. J. 31, 220–229 (2001).
Mayes, M. D. et al. Prevalence, incidence, survival, and disease characteristics of systemic sclerosis in a large US population. Arthritis Rheum. 48, 2246–2255 (2003).
De Martinis, M., Ciccarelli, F., Sirufo, M. M. & Ginaldi, L. An overview of environmental risk factors in systemic sclerosis. Expert Rev. Clin. Immunol. 12, 465–478 (2016).
McCormic, Z. D., Khuder, S. S., Aryal, B. K., Ames, A. L. & Khuder, S. A. Occupational silica exposure as a risk factor for scleroderma: a meta-analysis. Int. Arch. Occup. Environ. Health 83, 763–769 (2010).
Walczyk, M., Paradowska-Gorycka, A. & Olesinska, M. Epigenetics: the future direction in systemic sclerosis. Scand. J. Immunol. 86, 427–435 (2017).
Broen, J. C., Radstake, T. R. & Rossato, M. The role of genetics and epigenetics in the pathogenesis of systemic sclerosis. Nat. Rev. Rheumatol. 10, 671–681 (2014).
Englert, H. et al. Familial risk estimation in systemic sclerosis. Aust. N. Z. J. Med. 29, 36–41 (1999).
Arnett, F. C. et al. Familial occurrence frequencies and relative risks for systemic sclerosis (scleroderma) in three United States cohorts. Arthritis Rheum. 44, 1359–1362 (2001).
Feghali-Bostwick, C., Medsger, T. A. Jr & Wright, T. M. Analysis of systemic sclerosis in twins reveals low concordance for disease and high concordance for the presence of antinuclear antibodies. Arthritis Rheum. 48, 1956–1963 (2003).
Aho, K., Koskenvuo, M., Tuominen, J. & Kaprio, J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol. 13, 899–902 (1986).
Bammer, H., Schaltenbrand, G. & Solcher, H. [Examinations of twins in multiple sclerosis]. Dtsch. Z. Nervenheilkd 181, 261–279 (1960).
Selmi, C. et al. Primary biliary cirrhosis in monozygotic and dizygotic twins: genetics, epigenetics, and environment. Gastroenterology 127, 485–492 (2004).
Zhou, X., Tan, F. K., Xiong, M., Arnett, F. C. & Feghali-Bostwick, C. A. Monozygotic twins clinically discordant for scleroderma show concordance for fibroblast gene expression profiles. Arthritis Rheum. 52, 3305–3314 (2005).
Hudson, M. et al. Polyautoimmunity and familial autoimmunity in systemic sclerosis. J. Autoimmun. 31, 156–159 (2008).
Arora-Singh, R. K. et al. Autoimmune diseases and autoantibodies in the first degree relatives of patients with systemic sclerosis. J. Autoimmun. 35, 52–57 (2010).
Chairta, P., Nicolaou, P. & Christodoulou, K. Genomic and genetic studies of systemic sclerosis: a systematic review. Hum. Immunol. 78, 153–165 (2017).
Mayes, M. D. et al. Immunochip analysis identifies multiple susceptibility loci for systemic sclerosis. Am. J. Hum. Genet. 94, 47–61 (2014).
Arnett, F. C. et al. Major histocompatibility complex (MHC) class II alleles, haplotypes and epitopes which confer susceptibility or protection in systemic sclerosis: analyses in 1300 Caucasian, African-American and Hispanic cases and 1000 controls. Ann. Rheum. Dis. 69, 822–827 (2010).
Beretta, L. et al. Analysis of class II human leucocyte antigens in Italian and Spanish systemic sclerosis. Rheumatol. 51, 52–59 (2012).
Rodriguez-Reyna, T. S. et al. HLA class I and II blocks are associated to susceptibility, clinical subtypes and autoantibodies in Mexican systemic sclerosis (SSc) patients. PLoS ONE 10, e0126727 (2015).
Furukawa, H. et al. Human leukocyte antigen and systemic sclerosis in Japanese: the sign of the four independent protective alleles, DRB1*13:02, DRB1*14:06, DQB1*03:01, and DPB1*02:01. PLoS ONE 11, e0154255 (2016).
Wang, J. et al. Association of HLA-DPB1 with scleroderma and its clinical features in Chinese population. PLoS ONE 9, e87363 (2014).
Vigone, B. et al. Role of class II human leucocyte antigens in the progression from early to definite systemic sclerosis. Rheumatol. 54, 707–711 (2015).
Lambert, N. C. et al. HLA-DQA1*0501 is associated with diffuse systemic sclerosis in Caucasian men. Arthritis Rheum. 43, 2005–2010 (2000).
Stevens, A. M. et al. Brief report: HLA-DRB1, DQA1, and DQB1 in juvenile-onset systemic sclerosis. Arthritis Rheumatol. 68, 2772–2777 (2016).
Higgs, B. W. et al. Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann. Rheum. Dis. 70, 2029–2036 (2011).
Xu, Y., Wang, W., Tian, Y., Liu, J. & Yang, R. Polymorphisms in STAT4 and IRF5 increase the risk of systemic sclerosis: a meta-analysis. Int. J. Dermatol. 55, 408–416 (2016).
Lopez-Isac, E. et al. Brief report: IRF4 newly identified as a common susceptibility locus for systemic sclerosis and rheumatoid arthritis in a cross-disease meta-analysis of genome-wide association studies. Arthritis Rheumatol. 68, 2338–2344 (2016).
Allanore, Y. et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 7, e1002091 (2011).
Martin, J. E. et al. Identification of CSK as a systemic sclerosis genetic risk factor through genome wide association study follow-up. Hum. Mol. Genet. 21, 2825–2835 (2012).
Radstake, T. R. et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat. Genet. 42, 426–429 (2010).
Wang, J. et al. Lack of association of the CD247 SNP rs2056626 with systemic sclerosis in Han Chinese. Open Rheumatol. J. 8, 43–45 (2014).
Sato, S. et al. Levels of interleukin 12, a cytokine of type 1 helper T cells, are elevated in sera from patients with systemic sclerosis. J. Rheumatol. 27, 2838–2842 (2000).
Bossini-Castillo, L. et al. A GWAS follow-up study reveals the association of the IL12RB2 gene with systemic sclerosis in Caucasian populations. Hum. Mol. Genet. 21, 926–933 (2012).
Lopez-Isac, E. et al. Identification of IL12RB1 as a novel systemic sclerosis susceptibility locus. Arthritis Rheumatol. 66, 3521–3523 (2014).
Martin, J. E. et al. A systemic sclerosis and systemic lupus erythematosus pan-meta-GWAS reveals new shared susceptibility loci. Hum. Mol. Genet. 22, 4021–4029 (2013).
Tsuchiya, N. et al. Association of STAT4 polymorphism with systemic sclerosis in a Japanese population. Ann. Rheum. Dis. 68, 1375–1376 (2009).
Elshazli, R. & Settin, A. Association of PTPN22 rs2476601 and STAT4 rs7574865 polymorphisms with rheumatoid arthritis: a meta-analysis update. Immunobiology 220, 1012–1024 (2015).
Gestermann, N. et al. STAT4 is a confirmed genetic risk factor for Sjögren’s syndrome and could be involved in type 1 interferon pathway signaling. Genes Immun. 11, 432–438 (2010).
Namjou, B. et al. High-density genotyping of STAT4 reveals multiple haplotypic associations with systemic lupus erythematosus in different racial groups. Arthritis Rheum. 60, 1085–1095 (2009).
Tsuchiya, N., Ito, I. & Kawasaki, A. Association of IRF5, STAT4 and BLK with systemic lupus erythematosus and other rheumatic diseases. Nihon Rinsho Meneki Gakkai Kaishi 33, 57–65 (2010).
Ito, I. et al. Association of a functional polymorphism in the IRF5 region with systemic sclerosis in a Japanese population. Arthritis Rheum. 60, 1845–1850 (2009).
Koumakis, E. et al. A candidate gene study identifies a haplotype of CD2 as novel susceptibility factor for systemic sclerosis. Clin. Exp. Rheumatol. 34 (Suppl. 100), 43–48 (2016).
Sisirak, V. et al. Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell 166, 88–101 (2016).
Zochling, J. et al. An Immunochip-based interrogation of scleroderma susceptibility variants identifies a novel association at DNASE1L3. Arthritis Res. Ther. 16, 438 (2014).
Wei, P. et al. Identification of an association of TNFAIP3 polymorphisms with matrix metalloproteinase expression in fibroblasts in an integrative study of systemic sclerosis-associated genetic and environmental factors. Arthritis Rheumatol. 68, 749–760 (2016).
Zhao, J. H. et al. The influence of different solvents on systemic sclerosis: an updated meta-analysis of 14 case-control studies. J. Clin. Rheumatol. 22, 253–259 (2016).
Johar, A. S. et al. Candidate gene discovery in autoimmunity by using extreme phenotypes, next generation sequencing and whole exome capture. Autoimmun Rev. 14, 204–209 (2015).
Gao, L. et al. Identification of rare variants in ATP8B4 as a risk factor for systemic sclerosis by whole-exome sequencing. Arthritis Rheumatol. 68, 191–200 (2016).
Lopez-Isac, E. et al. Analysis of ATP8B4 F436L missense variant in a large systemic sclerosis cohort. Arthritis Rheumatol. 69, 1337–1338 (2017).
Mak, A. C. et al. Brief report: whole-exome sequencing for identification of potential causal variants for diffuse cutaneous systemic sclerosis. Arthritis Rheumatol. 68, 2257–2262 (2016).
Dieude, P. et al. BANK1 is a genetic risk factor for diffuse cutaneous systemic sclerosis and has additive effects with IRF5 and STAT4. Arthritis Rheum. 60, 3447–3454 (2009).
van Bon, L. et al. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. N. Engl. J. Med. 370, 433–443 (2014).
van Bon, L. et al. Distinct evolution of TLR-mediated dendritic cell cytokine secretion in patients with limited and diffuse cutaneous systemic sclerosis. Ann. Rheum. Dis. 69, 1539–1547 (2010).
Ah Kioon, M. D. et al. Plasmacytoid dendritic cells promote systemic sclerosis with a key role for TLR8. Sci. Transl Med. 10, eaam8458 (2018).
Silva-Cardoso, S. C. et al. CXCL4 exposure potentiates TLR-driven polarization of human monocyte-derived dendritic cells and increases stimulation of T cells. J. Immunol. 199, 253–262 (2017).
Altorok, N., Tsou, P. S., Coit, P., Khanna, D. & Sawalha, A. H. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann. Rheum. Dis. 74, 1612–1620 (2015).
Hattori, M. et al. Global DNA hypomethylation and hypoxia-induced expression of the ten eleven translocation (TET) family, TET1, in scleroderma fibroblasts. Exp. Dermatol. 24, 841–846 (2015).
Sargent, J. L. et al. A TGFβ-responsive gene signature is associated with a subset of diffuse scleroderma with increased disease severity. J. Invest. Dermatol. 130, 694–705 (2010).
Dantas, A. T. et al. Reassessing the role of the active TGF-β1 as a biomarker in systemic sclerosis: association of serum levels with clinical manifestations. Dis. Markers 2016, 6064830 (2016).
Dees, C. et al. The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann. Rheum. Dis. 73, 1232–1239 (2014).
Noda, S. et al. Simultaneous downregulation of KLF5 and FLI1 is a key feature underlying systemic sclerosis. Nat. Commun. 5, 5797 (2014).
Zhang, Y. et al. Poly(ADP-ribose) polymerase-1 regulates fibroblast activation in systemic sclerosis. Ann. Rheum. Dis. 77, 744–751 (2018).
Bujor, A. M. et al. Ciprofloxacin has antifibrotic effects in scleroderma fibroblasts via downregulation of DNMT1 and upregulation of FLI1. Int. J. Mol. Med. 30, 1473–1480 (2012).
Lei, W. et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand. J. Rheumatol. 38, 369–374 (2009).
Qing, Y. et al. Quantitation and mapping of the epigenetic marker 5-hydroxymethylcytosine. Bioessays https://doi.org/10.1002/bies.201700010 (2017).
Zhang, S. et al. miR-30a as potential therapeutics by targeting TET1 through regulation of Drp-1 promoter hydroxymethylation in idiopathic pulmonary fibrosis. Int. J. Mol. Sci. 18, E633 (2017).
O’Reilly, S., Ciechomska, M., Fullard, N., Przyborski, S. & van Laar, J. M. IL-13 mediates collagen deposition via STAT6 and microRNA-135b: a role for epigenetics. Sci. Rep. 6, 25066 (2016).
He, Y., Tsou, P. S., Khanna, D. & Sawalha, A. H. Methyl-CpG-binding protein 2 mediates antifibrotic effects in scleroderma fibroblasts. Ann. Rheum. Dis. 77, 1208–1218 (2018).
Keerthisingam, C. B. et al. Cyclooxygenase-2 deficiency results in a loss of the anti-proliferative response to transforming growth factor-β in human fibrotic lung fibroblasts and promotes bleomycin-induced pulmonary fibrosis in mice. Am. J. Pathol. 158, 1411–1422 (2001).
Evans, I. C. et al. Epigenetic regulation of cyclooxygenase-2 by methylation of C8orf4 in pulmonary fibrosis. Clin. Sci. (Lond.) 130, 575–586 (2016).
Kim, J. et al. TC1(C8orf4) is a novel endothelial inflammatory regulator enhancing NF-κB activity. J. Immunol. 183, 3996–4002 (2009).
Matatiele, P., Tikly, M., Tarr, G. & Gulumian, M. DNA methylation similarities in genes of black South Africans with systemic lupus erythematosus and systemic sclerosis. J. Biomed. Sci. 22, 34 (2015).
Ding, W. et al. Genome-wide DNA methylation analysis in systemic sclerosis reveals hypomethylation of IFN-associated genes in CD4(+) and CD8(+) T cells. J. Invest. Dermatol. 138, 1069–1077 (2018).
Yoshizaki, A. et al. Cell adhesion molecules regulate fibrotic process via Th1/Th2/Th17 cell balance in a bleomycin-induced scleroderma model. J. Immunol. 185, 2502–2515 (2010).
Wang, Y. et al. Hypomethylation and overexpression of ITGAL (CD11a) in CD4(+) T cells in systemic sclerosis. Clin. Epigenet. 6, 25 (2014).
Yin, H. et al. Histone demethylase JMJD3 regulates CD11a expression through changes in histone H3K27 tri-methylation levels in CD4+ T cells of patients with systemic lupus erythematosus. Oncotarget 8, 48938–48947 (2017).
Absher, D. M. et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T cell populations. PLoS Genet. 9, e1003678 (2013).
Carmona, F. D. et al. Novel identification of the IRF7 region as an anticentromere autoantibody propensity locus in systemic sclerosis. Ann. Rheum. Dis. 71, 114–119 (2012).
Rezaei, R. et al. IRF7 gene expression profile and methylation of its promoter region in patients with systemic sclerosis. Int. J. Rheum. Dis. 20, 1551–1561 (2017).
Wang, Y. Y. et al. DNA hypermethylation of the forkhead box protein 3 (FOXP3) promoter in CD4+ T cells of patients with systemic sclerosis. Br. J. Dermatol. 171, 39–47 (2014).
Almanzar, G. et al. Disease manifestation and inflammatory activity as modulators of Th17/Treg balance and RORC/FoxP3 methylation in systemic sclerosis. Int. Arch. Allergy Immunol. 171, 141–154 (2016).
Tsou, P. S. et al. Histone deacetylase 5 is overexpressed in scleroderma endothelial cells and impairs angiogenesis via repression of proangiogenic factors. Arthritis Rheumatol. 68, 2975–2985 (2016).
Wang, Y., Fan, P. S. & Kahaleh, B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 54, 2271–2279 (2006).
Chu, H. et al. Sirtuin1 protects against systemic sclerosis-related pulmonary fibrosis by decreasing proinflammatory and profibrotic processes. Am. J. Respir. Cell. Mol. Biol. 58, 28–39 (2018).
Zerr, P. et al. Sirt1 regulates canonical TGF-β signalling to control fibroblast activation and tissue fibrosis. Ann. Rheum. Dis. 75, 226–233 (2016).
Wei, J. et al. The histone deacetylase sirtuin 1 is reduced in systemic sclerosis and abrogates fibrotic responses by targeting transforming growth factor β signaling. Arthritis Rheumatol. 67, 1323–1334 (2015).
Wyman, A. E. et al. Sirtuin 7 is decreased in pulmonary fibrosis and regulates the fibrotic phenotype of lung fibroblasts. Am. J. Physiol. Lung Cell. Mol. Physiol. 312, L945–L958 (2017).
Sosulski, M. L., Gongora, R., Feghali-Bostwick, C., Lasky, J. A. & Sanchez, C. G. Sirtuin 3 deregulation promotes pulmonary fibrosis. J. Gerontol. A Biol. Sci. Med. Sci. 72, 595–602 (2017).
Akamata, K. et al. SIRT3 is attenuated in systemic sclerosis skin and lungs, and its pharmacologic activation mitigates organ fibrosis. Oncotarget 7, 69321–69336 (2016).
Zhu, X. et al. SIRT1 ameliorates systemic sclerosis by targeting the mTOR pathway. J. Dermatol. Sci. 87, 149–158 (2017).
Wang, Q. et al. Overexpression of JMJD3 may contribute to demethylation of H3K27me3 in CD4+ T cells from patients with systemic sclerosis. Clin. Immunol. 161, 396–399 (2015).
Bergmann, C. et al. The histone demethylase Jumonji domain-containing protein 3 (JMJD3) regulates fibroblast activation in systemic sclerosis. Ann. Rheum. Dis. 77, 150–158 (2018).
Fan, Z. et al. MKL1 is an epigenetic modulator of TGF-β induced fibrogenesis. Biochim. Biophys. Acta 1849, 1219–1228 (2015).
Rossato, M. et al. Association of microRNA-618 expression with altered frequency and activation of plasmacytoid dendritic cells in patients with systemic sclerosis. Arthritis Rheumatol. 69, 1891–1902 (2017).
Ciechomska, M. et al. The role of microRNA-5196 in the pathogenesis of systemic sclerosis. Eur. J. Clin. Invest. 47, 555–564 (2017).
Ciechomska, M., O’Reilly, S., Suwara, M., Bogunia-Kubik, K. & van Laar, J. M. MiR-29a reduces TIMP-1 production by dermal fibroblasts via targeting TGF-β activated kinase 1 binding protein 1, implications for systemic sclerosis. PLoS ONE 9, e115596 (2014).
Jafarinejad-Farsangi, S. et al. MicroRNA-29a induces apoptosis via increasing the Bax:Bcl-2 ratio in dermal fibroblasts of patients with systemic sclerosis. Autoimmunity 48, 369–378 (2015).
Jafarinejad-Farsangi, S. et al. Inhibition of microRNA-21 induces apoptosis in dermal fibroblasts of patients with systemic sclerosis. Int. J. Dermatol. 55, 1259–1267 (2016).
Zhou, B. et al. MicroRNA-202-3p regulates scleroderma fibrosis by targeting matrix metalloproteinase 1. Biomed. Pharmacother. 87, 412–418 (2017).
Yan, Q., Chen, J., Li, W., Bao, C. & Fu, Q. Targeting miR-155 to treat experimental scleroderma. Sci. Rep. 6, 20314 (2016).
Christmann, R. B. et al. MiR-155 in the progression of lung fibrosis in systemic sclerosis. Arthritis Res. Ther. 18, 155 (2016).
Artlett, C. M., Sassi-Gaha, S., Hope, J. L., Feghali-Bostwick, C. A. & Katsikis, P. D. Mir-155 is overexpressed in systemic sclerosis fibroblasts and is required for NLRP3 inflammasome-mediated collagen synthesis during fibrosis. Arthritis Res. Ther. 19, 144 (2017).
Artlett, C. M. et al. The inflammasome activating caspase 1 mediates fibrosis and myofibroblast differentiation in systemic sclerosis. Arthritis Rheum. 63, 3563–3574 (2011).
Iwamoto, N. et al. Downregulation of miR-193b in systemic sclerosis regulates the proliferative vasculopathy by urokinase-type plasminogen activator expression. Ann. Rheum. Dis. 75, 303–310 (2016).
Weiland, M., Gao, X. H., Zhou, L. & Mi, Q. S. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 9, 850–859 (2012).
Steen, S. O. et al. The circulating cell-free microRNA profile in systemic sclerosis is distinct from both healthy controls and systemic lupus erythematosus. J. Rheumatol. 42, 214–221 (2015).
Wuttge, D. M. et al. Specific autoantibody profiles and disease subgroups correlate with circulating micro-RNA in systemic sclerosis. Rheumatol. 54, 2100–2107 (2015).
Chouri, E. et al. Serum microRNA screening and functional studies reveal miR-483-5p as a potential driver of fibrosis in systemic sclerosis. J. Autoimmun. 89, 162–170 (2018).
Nakamura, K. et al. Altered expression of CD63 and exosomes in scleroderma dermal fibroblasts. J. Dermatol. Sci. 84, 30–39 (2016).
Wermuth, P. J., Piera-Velazquez, S. & Jimenez, S. A. Exosomes isolated from serum of systemic sclerosis patients display alterations in their content of profibrotic and antifibrotic microRNA and induce a profibrotic phenotype in cultured normal dermal fibroblasts. Clin. Exp. Rheumatol. 35 (Suppl. 106), 21–30 (2017).
Messemaker, T. C. et al. Antisense long non-coding RNAs are deregulated in skin tissue of patients with systemic sclerosis. J. Invest. Dermatol. 138, 826–835 (2018).
Wang, Z. et al. Long non-coding RNA TSIX is upregulated in scleroderma dermal fibroblasts and controls collagen mRNA stabilization. Exp. Dermatol. 25, 131–136 (2016).
Angiolilli, C., Baeten, D. L., Radstake, T. R. & Reedquist, K. A. The acetyl code in rheumatoid arthritis and other rheumatic diseases. Epigenomics 9, 447–461 (2017).
O’Reilly, S. Epigenetic modulation as a therapy in systemic sclerosis. Rheumatol. https://doi.org/10.1093/rheumatology/key071 (2018).
Strickland, F. M. et al. Characterisation of an epigenetically altered CD4(+) CD28(+) KIR(+) T cell subset in autoimmune rheumatic diseases by multiparameter flow cytometry. Lupus Sci. Med. 3, e000147 (2016).
Richardson, B. Effect of an inhibitor of DNA methylation on T cells. II. 5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum. Immunol. 17, 456–470 (1986).
Svegliati, S. et al. Oxidative DNA damage induces the ATM-mediated transcriptional suppression of the Wnt inhibitor WIF-1 in systemic sclerosis and fibrosis. Sci. Signal 7, ra84 (2014).
Huber, L. C. et al. Trichostatin A prevents the accumulation of extracellular matrix in a mouse model of bleomycin-induced skin fibrosis. Arthritis Rheum. 56, 2755–2764 (2007).
Ota, C. et al. Histone deacetylase inhibitor restores surfactant protein-C expression in alveolar-epithelial type II cells and attenuates bleomycin-induced pulmonary fibrosis in vivo. Exp. Lung Res. 41, 422–434 (2015).
Hu, L. et al. Epigenetic regulation of interleukin 6 by histone acetylation in macrophages and its role in paraquat-induced pulmonary fibrosis. Front. Immunol. 7, 696 (2016).
Ghosh, A. K. et al. p300 is elevated in systemic sclerosis and its expression is positively regulated by TGF-β: epigenetic feed-forward amplification of fibrosis. J. Invest. Dermatol. 133, 1302–1310 (2013).
Smoliga, J. M., Vang, O. & Baur, J. A. Challenges of translating basic research into therapeutics: resveratrol as an example. J. Gerontol. A Biol. Sci. Med. Sci. 67, 158–167 (2012).
Xiao, X. et al. EZH2 enhances the differentiation of fibroblasts into myofibroblasts in idiopathic pulmonary fibrosis. Physiol. Rep. 4, e12915 (2016).
Zeybel, M. et al. A proof-of-concept for epigenetic therapy of tissue fibrosis: inhibition of liver fibrosis progression by 3-deazaneplanocin A. Mol. Ther. 25, 218–231 (2017).
Ciechomska, M. et al. Histone demethylation and toll-like receptor 8-dependent cross-talk in monocytes promotes transdifferentiation of fibroblasts in systemic sclerosis via Fra-2. Arthritis Rheumatol. 68, 1493–1504 (2016).
Kramer, M. et al. Inhibition of H3K27 histone trimethylation activates fibroblasts and induces fibrosis. Ann. Rheum. Dis. 72, 614–620 (2013).
Shi, J. & Vakoc, C. R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736 (2014).
Tang, X. et al. Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. Am. J. Pathol. 183, 470–479 (2013).
O’Reilly, S. MicroRNAs in fibrosis: opportunities and challenges. Arthritis Res. Ther. 18, 11 (2016).
Tanaka, S. et al. Alteration of circulating miRNAs in SSc: miR-30b regulates the expression of PDGF receptor beta. Rheumatol. 52, 1963–1972 (2013).
Gallant-Behm, C. L. et al. Regulation of ECM production and fibrosis by MRG-201, a mimic of microRNA miR-29. Wound Repair Regener. 24, A9 (2016).
Gallant-Behm, C. L. et al. Pharmacodynamic activity of a microRNA-29b mimic (MRG-201) in human skin incisions. J. Investigative Dermatol. 137, B4 (2017).
Kader, F. & Ghai, M. DNA methylation-based variation between human populations. Mol. Genet. Genom. 292, 5–35 (2017).
Christopher, A. F. et al. MicroRNA therapeutics: discovering novel targets and developing specific therapy. Perspect. Clin. Res. 7, 68–74 (2016).
Guinea-Viniegra, J. et al. Targeting miR-21 to treat psoriasis. Sci. Transl Med. 6, 225re1 (2014).
Xu, P., Hu, G., Luo, C. & Liang, Z. DNA methyltransferase inhibitors: an updated patent review (2012–2015). Expert Opin. Ther. Pat. 26, 1017–1030 (2016).
Vojinovic, J. et al. Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 63, 1452–1458 (2011).
King, T. E. Jr., Noble, P. W. & Bradford, W. Z. Treatments for idiopathic pulmonary fibrosis. N. Engl. J. Med. 371, 783–784 (2014).
Marcellin, P. et al. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a 5-year open-label follow-up study. Lancet 381, 468–475 (2013).
Liaw, Y. F. Reversal of cirrhosis: an achievable goal of hepatitis B antiviral therapy. J. Hepatol. 59, 880–881 (2013).
Zeisberg, M. & Kalluri, R. Cellular mechanisms of tissue fibrosis. 1. Common and organ-specific mechanisms associated with tissue fibrosis. Am. J. Physiol. Cell Physiol. 304, C216–C225 (2013).
Tang, L., Chen, B., Ma, B. & Nie, S. Association between IRF5 polymorphisms and autoimmune diseases: a meta-analysis. Genet. Mol. Res. 13, 4473–4485 (2014).
Wang, J. et al. Association of the IRF5 SNP rs2004640 with systemic sclerosis in Han Chinese. Int. J. Immunopathol. Pharmacol. 27, 635–638 (2014).
Zhao, W. et al. The status of pulmonary fibrosis in systemic sclerosis is associated with IRF5, STAT4, IRAK1, and CTGF polymorphisms. Rheumatol. Int. 37, 1303–1310 (2017).
Luo, H., Zhu, H., Zhou, B., Xiao, X. & Zuo, X. MicroRNA-130b regulates scleroderma fibrosis by targeting peroxisome proliferator-activated receptor gamma. Mod. Rheumatol. 25, 595–602 (2015).
Nakayama, W. et al. Dysregulated interleukin-23 signalling contributes to the increased collagen production in scleroderma fibroblasts via balancing microRNA expression. Rheumatol. 56, 145–155 (2017).
Neveu, W. A., Mills, S. T., Staitieh, B. S. & Sueblinvong, V. TGF-β1 epigenetically modifies Thy-1 expression in primary lung fibroblasts. Am. J. Physiol. Cell Physiol. 309, C616–C626 (2015).
Palumbo-Zerr, K. et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat. Med. 21, 150–158 (2015).
Urban, J. R. & King, B. Divalproex sodium: a potential therapy for scleroderma digital ulcers. JAAD Case Rep. 1, 44–45 (2015).
Hinchcliff, M. et al. Mycophenolate mofetil treatment of systemic sclerosis reduces myeloid cell numbers and attenuates the inflammatory gene signature in skin. J. Invest. Dermatol. 138, 1301–1310 (2018).
Namas, R. et al. Efficacy of mycophenolate mofetil and oral cyclophosphamide on skin thickness: post hoc analyses from two randomized placebo-controlled trials. Arthritis Care Res. (Hoboken) 70, 439–444 (2018).
Volkmann, E. R. et al. Mycophenolate mofetil versus placebo for systemic sclerosis-related interstitial lung disease: an analysis of scleroderma lung studies I and II. Arthritis Rheumatol. 69, 1451–1460 (2017).
Tashkin, D. P. et al. Mycophenolate mofetil versus oral cyclophosphamide in scleroderma-related interstitial lung disease (SLS II): a randomised controlled, double-blind, parallel group trial. Lancet Respir. Med. 4, 708–719 (2016).
Reviewer information
Nature Reviews Rheumatology thanks Y. Asano, M. Whitfield and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
C.A., W.M., M.v.d.K. and E.C. researched data for the article. C.A., W.M., M.v.d.K., E.C. and T.R.D.J.R. wrote the article. C.A., W.M., K.A.R. and T.R.D.J.R. made substantial contributions to discussion of content. All authors reviewed and/or edited the manuscript before submission. M.v.d.K. and E.C. contributed equally to the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- DNA methylation
-
An epigenetic change to DNA that conventionally creates a close, inactive chromatin state that results in transcriptional repression.
- Histone post-translational modifications
-
Chemical modifications to histone tails (such as acetylation and methylation) that influence the accessibility of the DNA to the transcription machinery, thereby allowing or repressing gene expression.
- Immunochip
-
Single nucleotide polymorphism microarrays designed to replicate and establish statistically significant genome-wide association study loci associated with autoimmune and inflammatory disorders.
- MicroRNA
-
A short non-coding RNA of 18–24 nucleotides in length that regulates gene expression by binding to its complementary sequence within a target mRNA.
- Long non-coding RNA
-
A long non-coding RNA ~200 nucleotides in length that functions as a molecular scaffold to regulate gene expression at the transcriptional, post-transcriptional and epigenetic levels.
Rights and permissions
About this article
Cite this article
Angiolilli, C., Marut, W., van der Kroef, M. et al. New insights into the genetics and epigenetics of systemic sclerosis. Nat Rev Rheumatol 14, 657–673 (2018). https://doi.org/10.1038/s41584-018-0099-0
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41584-018-0099-0
This article is cited by
-
Distinct genome-wide DNA methylation and gene expression signatures in classical monocytes from African American patients with systemic sclerosis
Clinical Epigenetics (2023)
-
Epigenetic regulation of B cells and its role in autoimmune pathogenesis
Cellular & Molecular Immunology (2022)
-
How does age determine the development of human immune-mediated arthritis?
Nature Reviews Rheumatology (2022)
-
Epigenetic Dysregulation in Autoimmune and Inflammatory Skin Diseases
Clinical Reviews in Allergy & Immunology (2022)
-
Complement component C4 structural variation and quantitative traits contribute to sex-biased vulnerability in systemic sclerosis
npj Genomic Medicine (2022)