Abstract
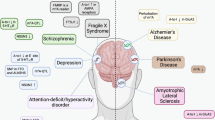
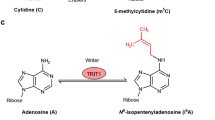
The field of epitranscriptomics examines the recently deciphered form of gene expression regulation that is mediated by type- and site-specific RNA modifications. Similarly to the role played by epigenetic mechanisms — which operate via DNA and histone modifications — epitranscriptomic modifications are involved in the control of the delicate gene expression patterns that are needed for the development and activity of the nervous system and are essential for basic and higher brain functions. Here we describe the mechanisms that are involved in the writing, erasing and reading of N6-methyladenosine, the most prevalent internal mRNA modification, and the emerging roles played by N6-methyladenosine in the nervous system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Goldberg, A. D., Allis, C. D. & Bernstein, E. Epigenetics: a landscape takes shape. Cell 128, 635–638 (2007).
Hwang, J.-Y., Aromolaran, K. A. & Zukin, R. S. The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 18, 347–361 (2017).
Yao, B. et al. Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 17, 537–549 (2016).
Dominissini, D. Roadmap to the epitranscriptome. Science 346, 1192–1192 (2014).
Li, X., Xiong, X. & Yi, C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2016).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). This article reports the first mapping and comprehensive characterization of the human and mouse m 6A methylomes. The work reported is also the first to discover m 6A-binding proteins belonging to the YTH domain-containing family.
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Dominissini, D. et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Li, X. et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316 (2016).
Levanon, E. Y. et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22, 1001–1005 (2004).
Squires, J. E. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012).
Delatte, B. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Li, X. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Akichika, S. et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II–associated methyltransferase. Science 363, eaav0080 (2019).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2016).
Sun, H., Zhang, M., Li, K., Bai, D. & Yi, C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 29, 80–82 (2019).
Arango, D. et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175, 1872–1886.e24 (2018).
Zhang, L.-S. et al. Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell 74, 1304–1316 (2019).
Shan, X., Tashiro, H. & Lin, C. L. The identification and characterization of oxidized RNAs in Alzheimer’s disease. J. Neurosci. 23, 4913–4921 (2003).
Dai, Q. et al. Nm-seq maps 2’-O-methylation sites in human mRNA with base precision. Nat. Methods 14, 695–698 (2017).
Dezi, V., Ivanov, C., Haussmann, I. U. & Soller, M. Nucleotide modifications in messenger RNA and their role in development and disease. Biochem. Soc. Trans. 44, 1385–1393 (2016).
Engel, M. & Chen, A. The emerging role of mRNA methylation in normal and pathological behavior. Genes Brain Behav. 17, e12428 (2018).
Jonkhout, N. et al. The RNA modification landscape in human disease. RNA 23, 1754–1769 (2017).
Kadumuri, R. V. & Janga, S. C. Epitranscriptomic code and its alterations in human disease. Trends Mol. Med. 24, 886–903 (2018).
Liu, E. Y., Cali, C. P. & Lee, E. B. RNA metabolism in neurodegenerative disease. Dis. Model. Mech. 10, 509–518 (2017).
Nussbacher, J. K., Batra, R., Lagier-Tourenne, C. & Yeo, G. W. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 38, 226–236 (2015).
Salta, E. & De Strooper, B. Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. 18, 627–640 (2017).
Noack, F. & Calegari, F. Epitranscriptomics: a new regulatory mechanism of brain development and function. Front. Neurosci. 12, 85 (2018).
Angelova, M. T. et al. The emerging field of epitranscriptomics in neurodevelopmental and neuronal disorders. Front. Bioeng. Biotechnol. 6, 46 (2018).
Leighton, L. J. et al. Experience-dependent neural plasticity, learning, and memory in the era of epitranscriptomics. Genes Brain Behav. 17, e12426 (2018).
Widagdo, J. & Anggono, V. The m6A-epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J. Neurochem. 147, 137–152 (2018).
Jung, Y. & Goldman, D. Role of RNA modifications in brain and behavior. Genes Brain Behav. 17, e12444 (2018).
Nachtergaele, S. & He, C. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 52, 349–372 (2018).
Geula, S. et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015). This study identifies METTL3 as a regulator for termination of murine naive pluripotency. It shows that m 6A on transcripts encoding pluripotency-promoting factors reduces their stability, thereby facilitating resolution of naive pluripotency and differentiation.
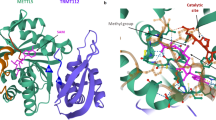
Śledź, P. & Jinek, M. J. E. Structural insights into the molecular mechanism of the m6A writer complex. Elife 5, e18434 (2016).
Wang, P., Doxtader, K. A. & Nam, Y. J. M. C. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Wang, X. et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578 (2016).
Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Schöller, E. et al. Interactions, localization, and phosphorylation of the m6A generating METTL3–METTL14–WTAP complex. RNA 24, 499–512 (2018).
Knuckles, P. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429 (2018).
Shi, H., Wei, J. & He, C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019).
Huang, H. et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567, 414–419 (2019).
Bertero, A. et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 555, 256–259 (2018).
Slobodin, B. et al. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169, 326–337 (2017).
Patil, D. P. et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). This study identifies and characterizes FTO as an m 6A demethylase, implying the dynamic nature of m 6A in mRNA.
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Wei, J. et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985 (2018).
Arguello, A. E., DeLiberto, A. N. & Kleiner, R. E. RNA chemical proteomics reveals the N6-methyladenosine (m6A)-regulated protein–RNA interactome. J. Am. Chem. Soc. 139, 17249–17252 (2017).
Edupuganti, R. R. et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878 (2017).
Zhao, B. S., Nachtergaele, S., Roundtree, I. A. & He, C. Our views of dynamic N6-methyladenosine RNA methylation. RNA 24, 268–272 (2018).
Stoilov, P., Rafalska, I. & Stamm, S. YTH: a new domain in nuclear proteins. Trends Biochem. Sci. 27, 495–497 (2002).
Kretschmer, J. et al. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′–3′ exoribonuclease XRN1. RNA 24, 1339–1350 (2018).
Roundtree, I. A. et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 6, e31311 (2017).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Shi, H. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Wang, X. et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Ries, R. J. et al. m6A enhances the phase separation potential of mRNA. Nature 571, 424–428 (2019). This article shows that polymethylated mRNA transcripts serve as a scaffold for binding of YTHDF1, YTHDF2, and YTHDF3, leading to liquid–liquid phase separation of YTHDF–mRNA complexes and partition into subcellular compartments such as P bodies, stress granules or neuronal RNA granules. It suggests that principles of phase separation further explain how m 6A governs RNA fate.
Huang, H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Wächter, K., Köhn, M., Stöhr, N. & Hüttelmaier, S. Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Biol. Chem. 394, 1077–1090 (2013).
Meyer, K. D. et al. 5′ UTR m6A promotes Cap-independent translation. Cell 163, 999–1010 (2015).
Roost, C. et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 137, 2107–2115 (2015).
Liu, N. et al. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564 (2015).
Liu, N. et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017).
Matsuki, H. et al. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cell 18, 135–146 (2013).
Tourrière, H. et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 (2003).
Zhou, K. I. et al. Regulation of co-transcriptional pre-mRNA splicing by m6A through the low-complexity protein hnRNPG. Mol. Cell 76, 70–81 (2019).
Haussmann, I. U. et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304 (2016).
Lence, T. et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247 (2016).
Ke, S. et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017).
Edens, B. M. et al. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 28, 845–854 (2019). This study shows a role for FMRP in mediating m 6A-dependent mRNA nuclear export through CRM1 during neural differentiation.
Nesterova, T. B. et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 10, 3129 (2019).
Xue, S. et al. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38 (2014).
Jackson, R. J., Hellen, C. U. T. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010).
Choi, J. et al. N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 23, 110–115 (2016).
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Park, O. H. et al. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 74, 494–507 (2019).
Batista, P. J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Hess, M. E. et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048 (2013).
Chang, M. et al. Region-specific RNA m6A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 7, 170166 (2017).
Ma, C. et al. RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19, 68 (2018).
Hu, W. F., Chahrour, M. H. & Walsh, C. A. The diverse genetic landscape of neurodevelopmental disorders. Annu. Rev. Genom. Hum. Genet. 15, 195–213 (2014).
Christian, K. M., Song, H. & Ming, G.-l. Functions and dysfunctions of adult hippocampal neurogenesis. Annu. Rev. Neurosci. 37, 243–262 (2014).
Atlasi, Y. & Stunnenberg, H. G. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 18, 643–658 (2017).
Weinberger, L., Ayyash, M., Novershtern, N. & Hanna, J. H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 17, 155–169 (2016).
Frye, M., Harada, B. T., Behm, M. & He, C. RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018).
Yoon, K.-J. et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171, 877–889 (2017).
Wang, Y. et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206 (2018).
Li, M. et al. Ythdf2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol. 19, 69 (2018).
Wu, R. et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29, 23–41 (2019).
Donega, V. et al. Transcriptional dysregulation in postnatal glutamatergic progenitors contributes to closure of the cortical neurogenic period. Cell Rep. 22, 2567–2574 (2018).
Stoeckli, E. T. Understanding axon guidance: are we nearly there yet? Development 145, dev151415 (2018).
Zhuang, M. et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 47, 4765–4777 (2019).
Wang, C.-X. et al. METTL3-mediated m6A modification is required for cerebellar development. PLOS Biol. 16, e2004880 (2018).
Gage, F. H. Mammalian neural stem cells. Science 287, 1433–1438 (2000).
Apple, D. M., Fonseca, R. S. & Kokovay, E. The role of adult neurogenesis in psychiatric and cognitive disorders. Brain Res. 1655, 270–276 (2017).
Kempermann, G. et al. Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell 23, 25–30 (2018).
Bond, Allison M., Ming, G.-l. & Song, H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell 17, 385–395 (2015).
Li, L. et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26, 2398–2411 (2017). The study finds a role for FTO in adult neurogenesis, learning and memory.
Vilar, M. & Mira, H. Regulation of neurogenesis by neurotrophins during adulthood: expected and unexpected roles. Front. Neurosci. 10, 26 (2016).
Glock, C., Heumüller, M. & Schuman, E. M. mRNA transport & local translation in neurons. Curr. Opin. Neurobiol. 45, 169–177 (2017).
Rangaraju, V., tom Dieck, S. & Schuman, E. M. Local translation in neuronal compartments: how local is local? EMBO Rep. 18, 693–711 (2017).
Van Driesche, S. J. & Martin, K. C. New frontiers in RNA transport and local translation in neurons. Dev. Neurobiol. 78, 331–339 (2018).
Zhang, F. et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27, 3936–3950 (2018).
Buffington, S. A., Huang, W. & Costa-Mattioli, M. Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 37, 17–38 (2014).
Hershey, J. W., Sonenberg, N. & Mathews, M. B. Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 4, a011528 (2012).
Peer, E., Moshitch-Moshkovitz, S., Rechavi, G. & Dominissini, D. The epitranscriptome in translation regulation. Cold Spring Harb. Perspect. Biol. 11, a032623 (2018).
Merkurjev, D. et al. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014 (2018). This study identifies synapse-specific m 6A methylation.
Perea, G., Navarrete, M. & Araque, A. Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 32, 421–431 (2009).
Shi, H. et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563, 249–253 (2018). This article shows that m 6A promotes protein translation of target transcripts in response to neuronal stimuli in the adult mouse hippocampus through YTHDF1 binding, thereby facilitating learning and memory.
Zhang, Z. et al. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 28, 1050–1061 (2018).
Banerjee, A., Ifrim, M. F., Valdez, A. N., Raj, N. & Bassell, G. J. Aberrant RNA translation in fragile X syndrome: from FMRP mechanisms to emerging therapeutic strategies. Brain Res. 1693, 24–36 (2018).
Dowling, R. J. O., Zakikhani, M., Fantus, I. G., Pollak, M. & Sonenberg, N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 67, 10804–10812 (2007).
Spaulding, E. L. & Burgess, R. W. Accumulating evidence for axonal translation in neuronal homeostasis. Front. Neurosci. 11, 312 (2017).
Cioni, J.-M., Koppers, M. & Holt, C. E. Molecular control of local translation in axon development and maintenance. Curr. Opin. Neurobiol. 51, 86–94 (2018).
Yu, J. et al. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46, 1412–1423 (2018). This study demonstrates m 6A-dependent regulation of axonal translation of methylated transcripts through axonally translated FTO. Non-nuclear, axonal FTO demethylates internal m 6A in axonal mRNA to control local translation affecting neuronal development.
Walters, B. J. et al. The role of the RNA demethylase FTO (fat mass and obesity-associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology 42, 1502–1510 (2017).
Donnelly, C. J. et al. Axonally synthesized β-actin and GAP-43 proteins support distinct modes of axonal growth. J. Neurosci. 33, 3311–3322 (2013).
Liu, X. et al. Genome‐wide association study of autism spectrum disorder in the East Asian populations. Autism Res. 9, 340–349 (2016).
Weng, Y.-L. et al. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325 (2018). This article shows that sciatic nerve lesion induces m 6A in transcripts of RAGs in the adult mouse dorsal root ganglion. YTHDF1 binding of m 6A-methylated transcripts promotes injury-induced protein translation and axon regeneration.
Costigan, M. et al. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 3, 16–34 (2002).
Mahar, M. & Cavalli, V. Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 19, 323–337 (2018).
Widagdo, J. et al. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36, 6771–6777 (2016).
Engel, M. et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron 99, 389–403 (2018).
Koranda, J. L. et al. Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99, 283–292 (2018).
Lerner, T. N. & Kreitzer, A. C. Neuromodulatory control of striatal plasticity and behavior. Curr. Opin. Neurobiol. 21, 322–327 (2011).
Acknowledgements
The authors thank the Kahn Family Foundation for continuous support of their research. D.D is supported by grants from the Israel Science Foundation (2494/18 and 2625/17) and the Human Frontier Science Program (CDA 00048/2018). G.R. and D.D are supported by the German–Israeli Project Cooperation (DIP) of the German Federal Ministry of Education and Research.
Author information
Authors and Affiliations
Contributions
D.D. and S.M-M. researched the data for the article, wrote and edited the manuscript before submission. G.R, N.A. and D.D. contributed substantially to discussions of the content. D.D., I.L., S.M.-M. and G.R. contributed to writing the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Epigenetic modifications
-
Covalent chemical modifications to DNA or histones that affect nucleosome positioning, DNA packaging and/or chromatin accessibility to provide spatial and temporal control of gene expression without altering the DNA sequence. These marks are stable and heritable, but can be removed by dedicated enzymes to provide dynamic regulation.
- Recognition elements
-
Chemically modified cis-acting elements that consist of specific nucleotide sequences or certain folded structures that can modulate the binding capacity of RNA-binding proteins.
- Internal mRNA modification
-
A chemical modification of the nucleotides in an mRNA transcript, excluding the 5′ cap structure.
- Phase separation
-
The process by which fibrils, hydrogels or liquid droplets are formed by low-complexity amino acid sequences. Physically, the process involves two solutes demixing to form two new phases of different composition.
- Processing bodies
-
(P bodies). Cytoplasmic membraneless RNA granules that dynamically store translationally inactive messenger ribonucleoproteins, similar to stress granules. P bodies can exist in the absence of stress and house proteins that promote mRNA decay and translational repression. Repressed messenger ribonucleoproteins can exit processing bodies and re-enter translation in the cytoplasm.
- Translation initiation factor
-
A protein that participates in the initiation stage of translation by guiding the recruitment, scanning and assembly of the 80S ribosome at the start codon of mRNA transcripts.
- Stress granules
-
Membraneless cytoplasmic aggregates of translationally inactive messenger ribonucleoproteins — composed of stalled preinitiation complexes, ribosomal subunits, translation initiation factors and mRNAs — that form in response to acute stress. Their formation and dissolution support proteome modulation during changing environmental conditions.
- Developmental competence
-
The ability of progenitors to appropriately respond to inductive differentiation cues towards specific lineages and follow the full spectrum of developmental trajectories: that is, to proliferate, differentiate and give rise to all possible progeny. During differentiation, stem cells commit to specific developmental pathways and lose competence to take on others.
- Pluripotency
-
A capacity of stem cells to differentiate into the three primary germ cell layers of the embryo (but not into extraembryonic tissues) and to self-renew by division.
- Induced pluripotent stem cell
-
A terminally differentiated somatic cell that has been reprogrammed back to an embryonic pluripotent stem cell-like state by the introduction of a set of pluripotency-related transcription factors in vitro. This technology provides an unlimited source of any cell for research and therapeutic applications.
- Organoids
-
Miniature, simplified three-dimensional versions of organs that are derived from stem cells.
- Long-term potentiation
-
A form of activity-dependent plasticity in which transient high-frequency stimulation produces a rapid and persistent increase in synaptic strength (transmission efficacy). Multiple forms of long-term potentiation exist, depending, among other variables, on the type of synapse, the nature of the stimulation and the developmental stage. It is thought to be a cellular mechanism underlying learning and memory.
- Fear conditioning
-
An associative form of learning in which pairing a neutral stimulus (as a particular context or cue) with an aversive event causes the former to acquire aversive properties and the ability to elicit fear responses when presented alone in a novel context (cued fear test) or by re-exposure to the conditioning context (contextual fear test).
Rights and permissions
About this article
Cite this article
Livneh, I., Moshitch-Moshkovitz, S., Amariglio, N. et al. The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci 21, 36–51 (2020). https://doi.org/10.1038/s41583-019-0244-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41583-019-0244-z
This article is cited by
-
The role of the methyltransferase METTL3 in prostate cancer: a potential therapeutic target
BMC Cancer (2024)
-
A neural m6A pathway regulates behavioral aggregation in migratory locusts
Science China Life Sciences (2024)
-
FTO-targeted siRNA delivery by MSC-derived exosomes synergistically alleviates dopaminergic neuronal death in Parkinson's disease via m6A-dependent regulation of ATM mRNA
Journal of Translational Medicine (2023)
-
The role of N-methyladenosine modification in acute and chronic kidney diseases
Molecular Medicine (2023)
-
Absolute quantification of single-base m6A methylation in the mammalian transcriptome using GLORI
Nature Biotechnology (2023)