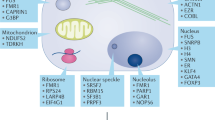
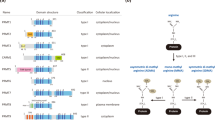
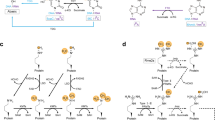
Abstract
Methylation of arginine residues by protein arginine methyltransferases (PRMTs) is involved in the regulation of fundamental cellular processes, including transcription, RNA processing, signal transduction cascades, the DNA damage response and liquid–liquid phase separation. Recent studies have provided considerable advances in the development of experimental tools and the identification of clinically relevant PRMT inhibitors. In this review, we discuss the regulation of PRMTs, their various cellular roles and the clinical relevance of PRMT inhibitors for the therapy of neurodegenerative diseases and cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
02 August 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Blanc, R. S. & Richard, S. Arginine methylation: the coming of age. Mol. Cell 65, 8–24 (2017).
Bedford, M. T. & Clarke, S. G. Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, (1–13 (2009).
Gayatri, S. & Bedford, M. T. Readers of histone methylarginine marks. Biochim. Biophys. Acta 1839, 702–710 (2014).
Akawi, N. et al. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat. Genet. 47, 1363–1369 (2015).
Agolini, E. et al. Expanding the clinical and molecular spectrum of PRMT7 mutations: 3 additional patients and review. Clin. Genet. 93, 675–681 (2018).
Nie, M. et al. CARM1-mediated methylation of protein arginine methyltransferase 5 represses human γ-globin gene expression in erythroleukemia cells. J. Biol. Chem. 293, 17454–17463 (2018).
Goulet, I., Gauvin, G., Boisvenue, S. & Côté, J. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J. Biol. Chem. 282, 33009–33021 (2007).
Baldwin, R. M., Morettin, A., Paris, G., Goulet, I. & Côté, J. Alternatively spliced protein arginine methyltransferase 1 isoform PRMT1v2 promotes the survival and invasiveness of breast cancer cells. Cell Cycle 11, 4597–4612 (2012).
Pollack, B. P. et al. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 274, 31531–31542 (1999).
Liu, F. et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell 19, 283–294 (2011).
Sipos, A. et al. Myosin phosphatase and RhoA-activated kinase modulate arginine methylation by the regulation of protein arginine methyltransferase 5 in hepatocellular carcinoma cells. Sci. Rep. 7, 40590 (2017).
Lattouf, H. et al. LKB1 regulates PRMT5 activity in breast cancer. Int. J. Cancer 144, 595–606 (2019).
Espejo, A. B. et al. PRMT5 C-terminal phosphorylation modulates a 14-3-3/PDZ interaction switch. J. Biol. Chem. 292, 2255–2265 (2017).
Guderian, G. & Peter, C. et al. RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. J. Biol. Chem. 286, 1976–1986 (2011).
Friesen, W. J. & Wyce, A. et al. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277, 8243–8247 (2002).
Aggarwal, P. et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18, 329–340 (2010).
Bao, X. et al. CSNK1a1 regulates PRMT1 to maintain the progenitor state in self-renewing somatic tissue. Dev. Cell 43, 227–239 (2017).
Chang, N. C. et al. The dystrophin glycoprotein complex regulates the epigenetic activation of muscle stem cell commitment. Cell Stem Cell 22, 755–768 (2018).
Zhang, H. T. et al. The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochem. Biophys. Acta 1863, 335–346 (2016).
Li, Z. et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat. Commun. 9, 1572 (2018).
Bhuripanyo, K. et al. Identifying the substrate proteins of U-box E3s E4B and CHIP by orthogonal ubiquitin transfer. Sci. Adv. 4, e1701393 (2018).
Li, X., Lai, Y., Li, J., Zou, M. & Zou, C. Oxidative stress destabilizes protein arginine methyltransferase 4 via glycogen synthase kinase 3β to impede lung epithelial cell migration. Am. J. Physiol. Cell Physiol. 313, C285–C294 (2017).
Shin, H. J. et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature 534, 553–557 (2016).
Lu, Y. F. et al. LncRNA SNHG16 functions as an oncogene by sponging miR-4518 and up-regulating PRMT5 expression in glioma. Cell. Physiol. Biochem. 45, 1975–1985 (2018).
Xu, Z. et al. MicroRNA-181 regulates CARM1 and histone arginine methylation to promote differentiation of human embryonic stem cells. PLOS ONE 8, e53146 (2013).
Vu, L. P. et al. PRMT4 blocks myeloid differentiation by assembling a methyl-RUNX1-dependent repressor complex. Cell Rep. 5, 1625–1638 (2013).
Li, B., Liu, L., Li, X. & Wu, L. miR-503 suppresses metastasis of hepatocellular carcinoma cell by targeting PRMT1. Biochem. Biophys. Res. Commun. 464, 982–987 (2015).
Zhang, H. T. et al. MiRNA-543 promotes osteosarcoma cell proliferation and glycolysis by partially suppressing PRMT9 and stabilizing HIF-1α protein. Oncotarget 8, 2342–2355 (2017).
Yang, Y. & Bedford, M. T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 (2013).
Huang, S., Litt, M. & Felsenfeld, G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 19, 1885–1893 (2005).
Zhao, X. et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 22, 640–653 (2008).
Shia, W. J. et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood 119, 4953–4962 (2012).
Cheung, N. et al. Targeting aberrant epigenetic networks mediated by PRMT1 and KDM4C in acute myeloid leukemia. Cancer Cell 29, 32–48 (2016).
Chen, D. et al. Regulation of transcription by a protein arginine methyltransferase. Science 284, 2174–2177 (1999).
Ceschin, D. G. et al. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 25, 1132–1146 (2011).
Yi, P. et al. Structural and functional impacts of ER coactivator sequential recruitment. Mol. Cell 67, 733–743 (2017).
Xu, W. et al. A transcriptional switch mediated by cofactor methylation. Science 294, 2507–2511 (2001).
Torres-Padilla, M. E., Parfitt, D. E., Kouzarides, T. & Zernicka-Goetz, M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218 (2007).
Hupalowska, A. et al. CARM1 and paraspeckles regulate pre-implantation mouse embryo development. Cell 175, 1902–1916 (2018). Arginine methylation is shown to have a role in paraspeckle regulation.
White, M. D. et al. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165, 75–87 (2016).
Hu, S. B. et al. Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. Genes Dev. 29, 630–645 (2015).
Pal, S., Vishwanath, S. N., Erdjument-Bromage, H., Tempst, P. & Sif, S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24, 9630–9645 (2004).
Scaglione, A. et al. PRMT5-mediated regulation of developmental myelination. Nat. Commun. 9, 2840 (2018).
Guccione, E. et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 (2007).
Waldmann, T. et al. Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 4, 11 (2011).
Iberg, A. N. et al. Arginine methylation of the histone H3 tail impedes effector binding. J. Biol. Chem. 283, 3006–3010 (2008).
Kirmizis, A. et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449, 928–932 (2007).
Migliori, V. et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 19, 136–144 (2012).
Chiang, K. et al. PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 21, 3498–3513 (2017).
Boisvert, F. M., Côté, J., Boulanger, M. C. & Richard, S. A. Proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics 2, 1319–1330 (2003).
Larsen, S. C. et al. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 9, rs9 (2016).
Musiani, D. et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal. 12, eaat8388 (2019).
Côté, J., Boisvert, F. M., Boulanger, M.-C., Bedford, M. T. & Richard, S. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell 14, 274–287 (2003).
Nichols, R. C. et al. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 256, 522–532 (2000).
Tradewell, M. L. et al. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum. Mol. Genet. 21, 136–149 (2012).
Wall, M. L. & Lewis, S. M. Methylarginines within the RGG-motif region of hnRNP A1 affect its IRES trans-acting factor activity and are required for hnRNP A1 stress granule localization and formation. J. Mol. Biol. 429, 295–307 (2017).
Murata, K. et al. PRMT1 deficiency in mouse juvenile heart induces dilated cardiomyopathy and reveals cryptic alternative splicing products. iScience 8, 200–213 (2018).
Meister, G. et al. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11, 1990–1994 (2001).
Meister, G. & Fischer, U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 21, 5853–5863 (2002).
Friesen, W. J., Massenet, S., Paushkin, S., Wyce, A. & Dreyfuss, G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7, 1111–1117 (2001).
Friesen, W. J. et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21, 8289–8300 (2001).
Zhang, Z. et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133, 585–600 (2008).
Yang, Y. et al. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 6, 6428 (2015). PRMT9 — a new type II PRMT — generates symmetrical dimethylarginine and regulates pre-mRNA splicing.
Hadjikyriacou, A., Yang, Y., Espejo, A., Bedford, M. T. & Clarke, S. G. Unique features of human protein arginine methyltransferase 9 (PRMT9) and its substrate RNA splicing factor SF3B2. J. Biol. Chem. 290, 16723–16743 (2015).
Cheng, D., Côté, J., Shaaban, S. & Bedford, M. T. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71–83 (2007).
Bezzi, M. et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 27, 1903–1916 (2013). PRMT5 is a key regulator of alternative splicing, influencing the p53 pathway.
Allende-Vega, N. et al. p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene 32, 1–14 (2013).
Koh, C. M. et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523, 96–100 (2015).
Dewaele, M. et al. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J. Clin. Invest. 126, 68–84 (2016).
Gerhart, S. V. et al. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci. Rep. 8, 9711 (2018).
Hamard, P. J. et al. PRMT5 regulates DNA repair by controlling the alternative splicing of histone-modifying enzymes. Cell Rep. 24, 2643–2657 (2018).
Inoue, M. et al. Arginine methylation controls the strength of γc-family cytokine signaling in T cell maintenance. Nat. Immunol. 19, 1265–1276 (2018).
Suganuma, T. et al. MPTAC determines APP fragmentation via sensing sulfur amino acid catabolism. Cell Rep. 24, 1585–1596 (2018).
Rengasamy, M. et al. The PRMT5/WDR77 complex regulates alternative splicing through ZNF326 in breast cancer. Nucleic Acids Res. 45, 11106–11120 (2017).
Close, P. et al. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature 484, 386–389 (2012).
Thandapani, P., O’Connor, T. R., Bailey, T. L. & Richard, S. Defining the RGG/RG motif. Mol. Cell 50, 613–623 (2013).
Roy, D. & Rajyaguru, P. I. Suppressor of clathrin deficiency (Scd6) — an emerging RGG-motif translation repressor. Wiley Interdiscip. Rev. RNA 22, e1479 (2018).
Poornima, G., Shah, S., Vignesh, V., Parker, R. & Rajyaguru, P. I. Arginine methylation promotes translation repression activity of eIF4G-binding protein, Scd6. Nucleic Acids Res. 44, 9358–9368 (2016).
Thandapani, P. et al. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. eLife 4, 06234 (2015).
Huang, L., Wang, Z., Narayanang, N. & Yang, Y. Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 46, 3061–3074 (2018).
Bachand, F. & Silver, P. A. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 23, 2641–2650 (2004).
Swiercz, R., Cheng, D., Kim, D. & Bedford, M. T. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J. Biol. Chem. 282, 16917–16923 (2007).
Dionne, K. L., Bergeron, D., Landry-Voyer, A.-M. & Bachand, F. The 40S ribosomal protein uS5 (RPS2) assembles into an extra-ribosomal complex with human ZNF277 that competes with the PRMT3-uS5 interaction. J. Biol. Chem. 294, 1944–1955 (2019).
Gao, G., Dhar, S. & Bedford, M. T. PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1. Nucleic Acids Res. 45, 4359–4369 (2017).
Ren, J. et al. Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J. Biol. Chem. 285, 12695–12705 (2010).
Raposo, A. E. & Piller, S. C. Protein arginine methylation: an emerging regulator of the cell cycle. Cell Div. 13, 3 (2018).
Yu, Z. et al. The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res. 22, 305–320 (2012).
Yuan, Q., Tian, R., Zhao, H., Li, L. & Bi, X. Multiple arginine residues are methylated in Drosophila Mre11 and required for survival following ionizing radiation. G3 8, 2099–2106 (2018).
Vadnais, C. et al. GFI1 facilitates efficient DNA repair by regulating PRMT1 dependent methylation of MRE11 and 53BP1. Nat. Commun. 9, 1418 (2018).
Polo, S. E. et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol. Cell 45, 505–516 (2012).
Gurunathan, G., Yu, Z., Coulombe, Y., Masson, J. Y. & Richard, S. Arginine methylation of hnRNPUL1 regulates interaction with NBS1 and recruitment to sites of DNA damage. Sci. Rep. 5, 10475 (2015).
Guendel, I. et al. Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function. PLOS ONE 5, e11379 (2010).
Boisvert, F.-M., Rhie, A., Richard, S. & Doherty, A. J. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 4, 1834–1841 (2005).
Adams, M. M. et al. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle 4, 1854–1861 (2005).
Pawlak, M. R., Scherer, C. A., Chen, J., Roshon, M. J. & Ruley, H. E. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. Biol. 20, 4859–4869 (2000).
Yu, Z., Chen, T., Hébert, J., Li, E. & Richard, S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol. Cell. Biol. 29, 2982–2996 (2009).
Fabbrizio, E. et al. Negatine regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3, 641–645 (2002).
Tee, W. W. et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 24, 2772–2777 (2010).
Huang, J., Vogel, G., Yu, Z., Almazan, G. & Richard, S. Type II arginine methyltransferase PRMT5 regulates gene expression of inhibitors of differentiation/DNA binding Id2 and Id4 during glial cell differentiation. J. Biol. Chem. 286, 44424–44432 (2011).
Clarke, T. L. et al. PRMT5-dependent methylation of the TIP60 coactivator RUVBL1 is a key regulator of homologous recombination. Mol. Cell 65, 900–916 (2017). Defines a role for PRMT5 in HR and the regulation of TIP60.
Tan, D. Q. et al. PRMT5 modulates splicing for genome integrity and preserves proteostasis of hematopoietic stem cells. Cell Rep. 26, 2316–2328 (2019).
Kaushik, S. et al. Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia 32, 499–509 (2018).
Guo, Z. et al. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat. Chem. Biol. 6, 766–773 (2010).
He, W. et al. A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res. 39, 4719–4727 (2011).
Rehman, I. et al. PRMT5-mediated arginine methylation of TDP1 for the repair of topoisomerase I covalent complexes. Nucleic Acids Res. 46, 5601–5617 (2018).
Aguilera, A. & Gomez-Gonzalez, B. DNA-RNA hybrids: the risks of DNA breakage during transcription. Nat. Struct. Mol. Biol. 24, 439–443 (2017).
Yang, Y. et al. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497 (2014).
Zhao, D. Y. et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 529, 48–53 (2016).
Mersaoui, S. et al. Arginine methylation of DDX5 RGG/RG motif by PRMT5 regulates RNA/DNA resolution. EMBO J. https://www.embopress.org/doi/10.15252/embj.2018100986.
Morales, J. C. et al. XRN2 links transcription termination to DNA damage and replication stress. PLOS Genet. 12, e1006107 (2016).
Skourti-Stathaki, K., Proudfoot, N. J. & Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011).
Xu, J. et al. Arginine methylation initiates BMP-induced Smad signaling. Mol. Cell 51, 5–19 (2013).
Katsuno, Y. et al. Arginine methylation of SMAD7 by PRMT1 in TGF-β-induced epithelial-mesenchymal transition and epithelial stem-cell generation. J. Biol. Chem. 293, 13059–13072 (2018).
Zhang, T. et al. Smad6 methylation represses NFκB activation and periodontal inflammation. J. Dent. Res. 97, 810–819 (2018).
Wei, H. et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc. Natl Acad. Sci. USA 110, 13516–13521 (2013).
Covic, M. et al. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-κB-dependent gene expression. EMBO J. 24, 85–96 (2005).
Hassa, P. O., Covic, M., Bedford, M. T. & Hottiger, M. O. Protein arginine methyltransferase 1 coactivates NF-κB-dependent gene expression synergistically with CARM1 and PARP1. J. Mol. Biol. 377, 668–678 (2008).
Gou, Y. et al. Protein arginine methyltransferase PRMT1 is essential for palatogenesis. J. Dent. Res. 97, 1510–1518 (2018).
Li, Q. et al. Histone arginine methylation by Prmt5 is required for lung branching morphogenesis through repression of BMP signaling. J. Cell Sci. 131, jcs217406 (2018).
Tamiya, H. et al. SHARPIN-mediated regulation of protein arginine methyltransferase 5 controls melanoma growth. J. Clin. Invest. 128, 517–530 (2018).
Tabata, T., Kokura, K., Ten Dijke, P. & Ishii, S. Ski co-repressor complexes maintain the basal repressed state of the TGF-β target gene, SMAD7, via HDAC3 and PRMT5. Genes Cells 14, 17–28 (2009).
Fu, T., Lv, X., Kong, Q. & Yuan, C. A novel SHARPIN-PRMT5-H3R2me1 axis is essential for lung cancer cell invasion. Oncotarget 8, 54809–54820 (2017).
Hsu, J. M. et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat. Cell Biol. 13, 174–181 (2011).
Calabretta, S. et al. Loss of PRMT5 promotes PDGFRα degradation during oligodendrocyte differentiation and myelination. Dev. Cell 46, 426–440 (2018).
Shashi, V. et al. The RBMX gene as a candidate for the Shashi X-linked intellectual disability syndrome. Clin. Genet. 88, 386–390 (2015).
Chong, P. A., Vernon, R. M. & Forman-Kay, J. D. RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665 (2018).
Vance, C. et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 (2009).
Kwiatkowski, T. J. J. et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009).
Patel, A. et al. A liquid-to- solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Qamar, S. et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734 (2018).
Hofweber, M. et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719 (2018). Arginine methylation suppresses RGG/RG-driven phase separation of the amyotrophic lateral sclerosis protein FUS.
Tanikawa, C. et al. Citrullination of RGG motifs in FET proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep. 22, 1473–1483 (2018).
Kim, H. J. et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013).
Ryan, V. H. et al. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465–479 (2018).
Gao, G. et al. PRMT1 loss sensitizes cells to PRMT5 inhibition. Nucleic Acids Res. 47, 5038–5048 (2019). PRMT1 loss was identified in a CRISPR–Cas9 screen as synthetic lethal with PRMT5 inhibition.
Fedoriw, A. et al. Anti-tumor activity of the first-in-class type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 through MTAP loss. Cancer Cell https://doi.org/10.1016/j.ccell.2019.05.014. MTAP-negative cancers are sensitive to the potent PRMT1 inhibitor GSK3368715.
Fong, J. Y. et al. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell https://doi.org/10.1016/j.ccell.2019.07.003 (2019). Inhibition of RNA splicing underlies the cytotoxic effects of PRMT inhibition.
Li, X., Wang, C., Jiang, H. & Luo, C. A patent review of arginine methyltransferase inhibitors (2010–2018). Expert Opin. Ther. Pat. 29, 97–114 (2019).
Cheng, D. et al. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 279, 23892–23899 (2004).
Feng, Y., Li, M., Wang, B. & Zheng, Y. G. Discovery and mechanistic study of a class of protein arginine methylation inhibitors. J. Med. Chem. 53, 6028–6039 (2010).
Eram, M. S. et al. A potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases. ACS Chem. Biol. 11, 772–781 (2016). Discovery and characterization of MS023 — a potent inhibitor of type I PRMTs.
Zhang, L. et al. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. eLife 4, e07938 (2015).
Jin, S., Mi, Y., Song, J., Zhang, P. & Liu, Y. PRMT1–RBM15 axis regulates megakaryocytic differentiation of human umbilical cord blood CD34+ cells. Exp. Ther. Med. 15, 2563–2568 (2018).
Siarheyeva, A. et al. An allosteric inhibitor of protein arginine methyltransferase 3. Structure 20, 1425–1435 (2012).
Kaniskan, H. Ü. et al. A potent, selective and cell-active allosteric inhibitor of protein arginine methyltransferase 3 (PRMT3). Angew. Chem. Int. Ed. 54, 5166–5170 (2015). Discovery and characterization of a potent, selective and cell-active allosteric inhibitor of PRMT3.
Mitchell, L. H. et al. Aryl pyrazoles as potent inhibitors of arginine methyltransferases: identification of the first PRMT6 tool compound. ACS Med. Chem. Lett. 6, 655–659 (2015).
Shen, Y. et al. Discovery of a potent, selective, and cell-active dual inhibitor of protein arginine methyltransferase 4 and protein arginine methyltransferase 6. J. Med. Chem. 59, 9124–9139 (2016).
Sack, J. S. et al. Structural basis for CARM1 inhibition by indole and pyrazole inhibitors. Biochem. J. 436, 331–339 (2011).
Nakayama, K. et al. TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget 9, 18480–18493 (2018).
Drew, A. E. et al. Identification of a CARM1 inhibitor with potent in vitro and in vivo activity in preclinical models of multiple myeloma. Sci. Rep. 7, 17993 (2017).
Greenblatt, S. M. et al. CARM1 is essential for myeloid leukemogenesis but dispensable for normal hematopoiesis. Cancer Cell 33, 1111–1127 (2018).
Liu, F. et al. PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nat. Cell Biol. 19, 1358–1370 (2017).
Wang, L. et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 30, 179–180 (2016).
Chan-Penebre, E. et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 11, 432–437 (2015).
Bonday, Z. Q. et al. LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med. Chem. Lett. 9, 612–617 (2018).
Brehmer, D. et al. DDT02-04: a novel PRMT5 inhibitor with potent in vitro and in vivo activity in preclinical lung cancer models. Cancer Res. 77 (Suppl.), DDT02-04 (2017).
Dvinge, H., Kim, E., Abdel-Wahab, O. & Bradley, R. K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16, 413–430 (2016).
Hsu, T. Y. et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525, 384–388 (2015).
Kryukov, G. V. et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351, 1214–1218 (2016).
Marjon, K. et al. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 15, 574–587 (2016).
Mavrakis, K. J. et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351, 1208–1213 (2016).
Dhar, S. et al. Loss of the major type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 3, 1311 (2013).
Szewczyk, M. M. et al. Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/503136v2 (2018).
Robak, P. & Robak, T. Bortezomib for the treatment of hematologic malignancies: 15 years later. Drugs R. D. 19, 73–92 (2019).
Milani, A., Basirnejad, M. & Bolhassani, A. Heat-shock proteins in diagnosis and treatment: an overview of different biochemical and immunological functions. Immunotherapy 11, 215–239 (2019).
Jain, K. & Clarke, S. G. PRMT7 as a unique member of the protein arginine methyltransferase family: a review. Arch. Biochem. Biophys. 665, 36–45 (2019).
Smart, A. C. et al. Intron retention is a source of neoepitopes in cancer. Nat. Biotechnol. 36, 1056–1058 (2018).
Nagai, Y. et al. PRMT5 associates with the FOXP3 homomer and when disabled enhances targeted p185erbB2/neu tumor immunotherapy. Front. Immunol. 10, 174 (2019).
Nguyen, H. D. et al. Spliceosome mutations induce R loop-associated sensitivity to ATR inhibition in myelodysplastic syndromes. Cancer Res. 78, 5363–5374 (2018).
Chen, L. et al. The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol. Cell 69, 412–425 (2018).
Nacev, B. A. et al. The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 567, 473–478 (2019).
Fontecave, M., Atta, M. & Mulliez, E. S-adenosylmethionine: nothing goes to waste. Trends Biochem. Sci. 29, 243–249 (2004).
Walport, L. J. et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974 (2016).
Li, S. et al. JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep. 23, 389–403 (2018).
Raijmakers, R. et al. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J. Mol. Biol. 367, 1118–1129 (2007).
Zhang, H., Chen, Z. H. & Savarese, T. M. Codeletion of the genes for p16 INK4, methylthioadenosine phosphorylase, interferon-α1, interferon-β1, and other 9p21 markers in human malignant cell lines. Cancer Genet. Cytogenet. 86, 22–28 (1996).
Liu, X., Xu, X., Shang, R. & Chen, Y. Asymmetric dimethylarginine (ADMA) as an important risk factor for the increased cardiovascular diseases and heart failure in chronic kidney disease. Nitric Oxide 78, 113–120 (2018).
Infantino, S., Benz, B., Waldmann, T., Jung, M., Schneider, R. & Reth, M. Arginine methylation of the B cell antigen receptor promotes differentiation. J. Exp. Med. 207, 711–719 (2010).
Geoghegan, V., Guo, A., Trudgian, D., Thomas, B. & Acuto, O. Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat. Commun. 6, 6758 (2015).
Infantino, S. et al. Arginine methylation catalyzed by PRMT1 is required for B cell activation and differentiation. Nat. Commun. 8, 891 (2017).
Litzler, L. C. et al. PRMT5 is essential for B cell development and germinal center dynamics. Nat. Commun. 10, 22 (2019).
Somasundaram, V. et al. Molecular mechanisms of nitric oxide in cancer progression, signal transduction, and metabolism. Antioxid. Redox Signal. 30, 1124–1143 (2019).
Rodriguez, P. C., Ochoa, A. C. & Al-Khami, A. A. Arginine metabolism in myeloid cells shapes innate and adaptive immunity. Front. Immunol. 8, 93 (2017).
Acknowledgements
E.G. acknowledges funding support from the NRF2016-CRP001-103 CRP award and from the RNA Biology Center at the Cancer Science Institute (CSI) of Singapore, National University of Singapore, and funding by the Singapore Ministry of Education’s Tier 3 grants (grant number MOE2014-T3-1-006). S.R. acknowledges funding from the Canadian Institutes of Health Research (grant number FDN-154303).
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to researching data for the article, the discussion of content, and writing and editing of the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Molecular Cell Biology thanks M. Bedford, C. Davies and S. Nimer for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- π-stacking interactions
-
Interactions between rings of aromatic amino acids.
- Aromatic cages
-
Refers to a protein structure that recognizes trimethyllysine (Me3+) ions.
- Paraspeckle
-
Membraneless nuclear compartment of poorly understood functions, which contains ribonucleoprotein particles.
- Small nuclear ribonucleoproteins
-
(snRNPs). Ribonucleoprotein complexes that function in pre-mRNA splicing; they are assembled in the cytoplasm and imported into the nucleus.
- Homologous recombination
-
(HR). A DNA double-strand break repair process, in which homologous sequences (usually sister chromatids) are used as templates for high fidelity repair.
- Stress granules
-
Protein and RNA cytoplasmic aggregations that appear in stress conditions and contain translationally repressed mRNAs.
- Internal ribosome entry site
-
(IRES). Internal mRNA sites that mediate cap-independent translation initiation.
- Non-homologous end joining
-
A DNA double-strand break repair process, in which the two break ends are re-joined without the use of a homologous template.
- Poly(ADP-ribose) polymerase
-
A family of enzymes that catalyse the addition of polymers of ADP-ribose to proteins.
- R-loops
-
RNA–DNA hybrids, in which a single-stranded RNA hybridizes to a template strand in a DNA duplex and displaces the non-template strand as a loop.
- Liquid–liquid phase separation
-
A process in which different liquids undergo demixing (phase separation) from their cellular surrounding, for example, in the formation of membraneless bodies.
- Non-competitive
-
Refers to inhibitors that are able to reduce enzymatic activity while binding to the targeted enzyme in the presence of S-adenosylmethionine (SAM-non-competitive) or the substrate (substrate-non-competitive).
Rights and permissions
About this article
Cite this article
Guccione, E., Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol 20, 642–657 (2019). https://doi.org/10.1038/s41580-019-0155-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-019-0155-x
This article is cited by
-
PRMT2 silencing regulates macrophage polarization through activation of STAT1 or inhibition of STAT6
BMC Immunology (2024)
-
Current and future therapeutic strategies for high-grade gliomas leveraging the interplay between epigenetic regulators and kinase signaling networks
Journal of Experimental & Clinical Cancer Research (2024)
-
Epigenetic modifications in the development of bronchopulmonary dysplasia: a review
Pediatric Research (2024)
-
GlPRMT5 inhibits GlPP2C1 via symmetric dimethylation and regulates the biosynthesis of secondary metabolites in Ganoderma lucidum
Communications Biology (2024)
-
Loss-of-function mutation in PRMT9 causes abnormal synapse development by dysregulation of RNA alternative splicing
Nature Communications (2024)