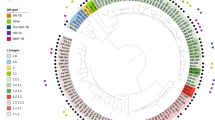
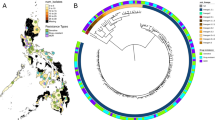
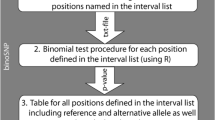
Abstract
Whole genome sequencing (WGS) of Mycobacterium tuberculosis has rapidly progressed from a research tool to a clinical application for the diagnosis and management of tuberculosis and in public health surveillance. This development has been facilitated by drastic drops in cost, advances in technology and concerted efforts to translate sequencing data into actionable information. There is, however, a risk that, in the absence of a consensus and international standards, the widespread use of WGS technology may result in data and processes that lack harmonization, comparability and validation. In this Review, we outline the current landscape of WGS pipelines and applications, and set out best practices for M. tuberculosis WGS, including standards for bioinformatics pipelines, curated repositories of resistance-causing variants, phylogenetic analyses, quality control and standardized reporting.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
World Health Organization. Global tuberculosis report 2018. WHO https://www.who.int/tb/publications/global_report/archive/ (2018).
The CRyPTIC Consortium and the 100,000 Genomes Project. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N. Engl. J. Med. 379, 1403–1415 (2018).The first large-scale study demonstrating how phenotypic testing can be replaced by WGS for first-line drug testing.
Gardy, J. L. et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 364, 730–739 (2011).
Cole, S. T. et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998).
Cabibbe, A. M., Walker, T. M., Niemann, S. & Cirillo, D. M. Whole genome sequencing of Mycobacterium tuberculosis. Eur. Respir. J. 52, 1801163 (2018).
Satta, G. et al. Mycobacterium tuberculosis and whole-genome sequencing: how close are we to unleashing its full potential? Clin. Microbiol. Infect. 24, 604–609 (2018).An extensive review of the literature outlining the potential of WGS for TB research and clinical use.
Lipworth, S. et al. SNP-IT tool for identifying subspecies and associated lineages of Mycobacterium tuberculosis complex. Emerg. Infect. Dis. 25, 482–488 (2019).
Coll, F. et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat. Commun. 5, 4812 (2014).This study reports the now standard sublineage typing scheme using SNP-based information for MTBC.
Homolka, S. et al. High resolution discrimination of clinical Mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLOS ONE 7, e39855 (2012).
Trauner, A. et al. The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biol. 18, 71 (2017).
Merker, M., Kohl, T. A., Niemann, S. & Supply, P. The evolution of strain typing in the Mycobacterium tuberculosis complex. Adv. Exp. Med. Biol. 1019, 43–78 (2017).
Jajou, R. et al. Epidemiological links between tuberculosis cases identified twice as efficiently by whole genome sequencing than conventional molecular typing: A population-based study. PLOS ONE 13, e0195413 (2018). This study shows the advantage of WGS approaches over mycobacterial interspersed repetitive unit variable-number tandem repeat genotyping for detection of transmission clusters.
Wyllie, D. H. et al. A quantitative evaluation of MIRU-VNTR typing against whole-genome sequencing for identifying Mycobacterium tuberculosis transmission: a prospective observational cohort study. EBioMedicine 34, 122–130 (2018).
Walker, T. M. et al. A cluster of multidrug-resistant Mycobacterium tuberculosis among patients arriving in Europe from the Horn of Africa: a molecular epidemiological study. Lancet Infect. Dis. 18, 431–440 (2018).
Tagliani, E. et al. EUSeqMyTB to set standards and build capacity for whole genome sequencing for tuberculosis in the EU. Lancet Infect. Dis. 18, 377 (2018). Announcement of the European Centre for Disease Prevention and Control efforts to establish and validate the use of WGS for all TB public health initiatives.
Cohen, K. A. et al. Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLOS Med. 12, e1001880 (2015).
Eldholm, V. et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat. Commun. 6, 7119 (2015).
Merker, M. et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat. Genet. 47, 242–249 (2015).
Zignol, M. et al. Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study. Lancet Infect. Dis. 18, 675–683 (2018).
Gröschel, M. I. et al. Pathogen-based precision medicine for drug-resistant tuberculosis. PLOS Pathog. 14, e1007297 (2018).
World Health Organization. The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex: technical guide. WHO https://apps.who.int/iris/handle/10665/274443 (2018). This guide is the first step towards validation of WGS as a tool for MTBC clinical and public health work.
Nebenzahl-Guimaraes, H., Jacobson, K. R., Farhat, M. R. & Murray, M. B. Systematic review of allelic exchange experiments aimed at identifying mutations that confer drug resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 69, 331–342 (2014).
Sandgren, A. et al. Tuberculosis drug resistance mutation database. PLOS Med. 6, e1000002 (2009).
Coll, F. et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 7, 51 (2015).
Miotto, P. et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur. Respir. J. 50, 1701354 (2017). This was the first study to use a score system to classify mutations for clinical interpretation.
Starks, A. M. et al. Collaborative effort for a centralized worldwide tuberculosis relational sequencing data platform. Clin. Infect. Dis. 61, S141–S146 (2015). This publication outlines the design and use of the ReSeqTB platform.
Brown, T., Nikolayevskyy, V., Velji, P. & Drobniewski, F. Associations between Mycobacterium tuberculosis strains and phenotypes. Emerg. Infect. Dis. 16, 272–280 (2010).
Comas, I. et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45, 1176–1182 (2013).
Meehan, C. J. et al. The relationship between transmission time and clustering methods in Mycobacterium tuberculosis epidemiology. EBioMedicine 37, 410–416 (2018).
Kohl, T. A. et al. Harmonized genome wide typing of tubercle bacilli using a web-based gene-by-gene nomenclature system. EBioMedicine 34, 131–138 (2018).
Walker, T. M. et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13, 137–146 (2013).
Koster, K. J. et al. Genomic sequencing is required for identification of tuberculosis transmission in Hawaii. BMC Infect. Dis. 18, 608 (2018).
Kohl, T. A. et al. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 6, e5895 (2018).
Ezewudo, M. et al. Integrating standardized whole genome sequence analysis with a global Mycobacterium tuberculosis antibiotic resistance knowledgebase. Sci. Rep. 8, 15382 (2018).
Brynildsrud, O. B. et al. Global expansion of Mycobacterium tuberculosis lineage 4 shaped by colonial migration and local adaptation. Sci. Adv. 4, eaat5869 (2018).
Brown, A. C. et al. Rapid whole-genome sequencing of Mycobacterium tuberculosis isolates directly from clinical samples. J. Clin. Microbiol. 53, 2230–2237 (2015).
Conceição, E. C. et al. Analysis of potential household transmission events of tuberculosis in the city of Belem, Brazil. Tuberculosis 113, 125–129 (2018).
Walker, T. M. et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect. Dis. 15, 1193–1202 (2015).
Goig, G. A., Blanco, S., Garcia-Basteiro, A. & Comas, I. Pervasive contaminations in sequencing experiments are a major source of false genetic variability: a Mycobacterium tuberculosis meta-analysis. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/403824v1 (2018).
Menardo, F. et al. Treemmer: a tool to reduce large phylogenetic datasets with minimal loss of diversity. BMC Bioinformatics 19, 164 (2018).
Bryant, J. M. et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect. Dis. 13, 110 (2013).
Shea, J. et al. Comprehensive whole-genome sequencing and reporting of drug resistance profiles on clinical cases of Mycobacterium tuberculosis in New York state. J. Clin. Microbiol. 55, 1871–1882 (2017).
Phelan, J. et al. Mycobacterium tuberculosis whole genome sequencing and protein structure modelling provides insights into anti-tuberculosis drug resistance. BMC Med. 14, 31 (2016).
Witney, A. A. et al. Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial. BMC Med. 15, 71 (2017).
Casali, N. et al. Whole genome sequence analysis of a large isoniazid-resistant tuberculosis outbreak in London: a retrospective observational study. PLOS Med. 13, e1002137 (2016).
Feuerriegel, S. et al. PhyResSE: a web tool delineating Mycobacterium tuberculosis antibiotic resistance and lineage from whole-genome sequencing data. J. Clin. Microbiol. 53, 1908–1914 (2015).
Bradley, P. et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun. 6, 10063 (2015).
Iwai, H., Kato-Miyazawa, M., Kirikae, T. & Miyoshi-Akiyama, T. CASTB (the comprehensive analysis server for the Mycobacterium tuberculosis complex): a publicly accessible web server for epidemiological analyses, drug-resistance prediction and phylogenetic comparison of clinical isolates. Tuberculosis 95, 843–844 (2015).
Steiner, A., Stucki, D., Coscolla, M., Borrell, S. & Gagneux, S. KvarQ: targeted and direct variant calling from fastq reads of bacterial genomes. BMC Genomics 15, 881 (2014).
Farhat, M. et al. genTB: translational genomics of tuberculosis. genTB https://gentb.hms.harvard.edu (2015).
Schleusener, V., Köser, C. U., Beckert, P., Niemann, S. & Feuerriegel, S. Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools. Sci. Rep. 7, 46327 (2017).
Ngo, T.-M. & Teo, Y.-Y. Genomic prediction of tuberculosis drug-resistance: benchmarking existing databases and prediction algorithms. BMC Bioinformatics 20, 68 (2019).
Phelan, J. et al. The variability and reproducibility of whole genome sequencing technology for detecting resistance to anti-tuberculous drugs. Genome Med. 8, 132 (2016).
Macedo, R. et al. Dissecting whole-genome sequencing-based online tools for predicting resistance in Mycobacterium tuberculosis: can we use them for clinical decision guidance? Tuberculosis 110, 44–51 (2018).
Angers-Loustau, A. et al. The challenges of designing a benchmark strategy for bioinformatics pipelines in the identification of antimicrobial resistance determinants using next generation sequencing technologies. F1000Res. 7, 459 (2018).
US Food and Drug Administration. Infectious disease next generation sequencing based diagnostic devices: microbial identification and detection of antimicrobial resistance and virulence markers. FederalRegister.gov https://www.federalregister.gov/documents/2016/08/11/2016-19109/infectious-disease-next-generation-sequencing-based-diagnostic-devices-microbial-identification-and (2016).
Pouseele, H. & Supply, P. Accurate whole-genome sequencing-based epidemiological surveillance of Mycobacterium tuberculosis. Methods Microbiol. 42, 359–394 (2015).
Simonyan, V., Goecks, J. & Mazumder, R. Biocompute objects — a step towards evaluation and validation of biomedical scientific computations. PDA J. Pharm. Sci. Technol. 71, 136–146 (2017).
Alterovitz, G. et al. Enabling precision medicine via standard communication of HTS provenance, analysis, and results. PLOS Biol. 16, e3000099 (2018).
Stucki, D. et al. Standard genotyping overestimates transmission of Mycobacterium tuberculosis among immigrants in a low-incidence country. J. Clin. Microbiol. 54, 1862–1870 (2016).
Liu, Q. et al. China’s tuberculosis epidemic stems from historical expansion of four strains of Mycobacterium tuberculosis. Nat. Ecol. Evol. 2, 1982–1992 (2018).
Holt, K. E. et al. Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat. Genet. 50, 849–856 (2018).
Coll, F. et al. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat. Genet. 50, 307–316 (2018).
Farhat, M. R. et al. GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nat. Commun. https://doi.org/10.1038/s41467-019-10110-6 (2019).
Kwong, J. C., Mccallum, N., Sintchenko, V. & Howden, B. P. Whole genome sequencing in clinical and public health microbiology. Pathology 47, 199–210 (2015).
Crisan, A., McKee, G., Munzner, T. & Gardy, J. L. Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory. PeerJ 6, e4218 (2017). This article reports new standards for reporting of WGS-based TB clinical information.
Tornheim, J. A. et al. Building the framework for standardized clinical laboratory reporting of next generation sequencing data for resistance-associated mutations in Mycobacterium tuberculosis complex. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciz219 (2019).
Tan, T. W. et al. Advancing standards for bioinformatics activities: persistence, reproducibility, disambiguation and Minimum Information About a Bioinformatics investigation (MIABi). BMC Genomics 11 (Suppl. 4), 27 (2010).
Field, N. et al. Strengthening the Reporting of Molecular Epidemiology for Infectious Diseases (STROME-ID): an extension of the STROBE statement. Lancet Infect. Dis. 14, 341–352 (2014).
World Health Organization. WHO’s code of conduct for open and timely sharing of pathogen genetic sequence data during outbreaks of infectious disease. WHO https://www.who.int/blueprint/what/norms-standards/gsdsharing/en/ (2019).
Allard, M. W. et al. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol. 54, 1975–1983 (2016).
Karikari, T. K. Bioinformatics in Africa: the rise of Ghana? PLOS Comput. Biol. 11, e1004308 (2015).
Tekola-Ayele, F. & Rotimi, C. N. Translational genomics in low- and middle-income countries: opportunities and challenges. Public Health Genomics 18, 242–247 (2015).
Helmy, M., Awad, M. & Mosa, K. A. Limited resources of genome sequencing in developing countries: challenges and solutions. Appl. Transl Genom. 9, 15–19 (2016).
Satta, G., Atzeni, A. & McHugh, T. D. Mycobacterium tuberculosis and whole genome sequencing: a practical guide and online tools available for the clinical microbiologist. Clin. Microbiol. Infect. 23, 69–72 (2017).
Kurtzer, G. M., Sochat, V. & Bauer, M. W. Singularity: scientific containers for mobility of compute. PLOS ONE 12, e0177459 (2017).
Merkel, D. Docker: lightweight Linux containers for consistent development and deployment. Linux J. 2014, 2 (2014).
Grüning, B. et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods 15, 475–476 (2018).
Jackman, S., Birol, I., Jackman, S. & Birol, I. Linuxbrew and Homebrew for cross-platform package management. F1000Res. 5, 1795 (2016).
Langille, M. G. I. & Eisen, J. A. BioTorrents: a file sharing service for scientific data. PLOS ONE 5, e10071 (2010).
Karikari, T. K., Quansah, E. & Mohamed, W. M. Y. Widening participation would be key in enhancing bioinformatics and genomics research in Africa. Appl. Transl Genom. 6, 35–41 (2015).
Bah, S. Y., Morang’a, C. M., Kengne-Ouafo, J. A., Amenga–Etego, L. & Awandare, G. A. Highlights on the application of genomics and bioinformatics in the fight against infectious diseases: challenges and opportunities in Africa. Front. Genet. 9, 575 (2018).
Zignol, M. et al. Population-based resistance of Mycobacterium tuberculosis isolates to pyrazinamide and fluoroquinolones: results from a multicountry surveillance project. Lancet Infect. Dis. 16, 1185–1192 (2016).
Kumwenda, S. et al. Challenges facing young African scientists in their research careers: a qualitative exploratory study. Malawi Med. J. 29, 1–4 (2017).
Rabbani, F. et al. Schools of public health in low and middle-income countries: an imperative investment for improving the health of populations? BMC Public Health 16, 941 (2016).
Helb, D. et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48, 229–237 (2010).
Wyllie, D. H. et al. Control of artifactual variation in reported intersample relatedness during clinical use of a Mycobacterium tuberculosis sequencing pipeline. J. Clin. Microbiol. 56, e00104–18 (2018).
Wood, D. E. & Salzberg, S. L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014).
Kim, D., Song, L., Breitwieser, F. P. & Salzberg, S. L. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 26, 1721–1729 (2016).
Médigue, C., Cole, S. T., Camus, J.-C. & Pryor, M. J. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 148, 2967–2973 (2002).
Periwal, V. et al. Comparative whole-genome analysis of clinical isolates reveals characteristic architecture of Mycobacterium tuberculosis pangenome. PLOS ONE 10, e0122979 (2015).
Gao, Q. et al. Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology 151, 5–14 (2005).
Kato-Maeda, M. et al. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11, 547–554 (2001).
Alland, D. et al. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J. Clin. Microbiol. 45, 39–46 (2007).
Ioerger, T. R. et al. Variation among genome sequences of H37Rv strains of Mycobacterium tuberculosis from multiple laboratories. J. Bacteriol. 192, 3645–3653 (2010).
Lee, R. S. & Behr, M. A. Does choice matter? Reference-based alignment for molecular epidemiology of tuberculosis. J. Clin. Microbiol. 54, 1891–1895 (2016).
Norman, A., Folkvardsen, D. B., Overballe-Petersen, S. & Lillebaek, T. Complete genome sequence of Mycobacterium tuberculosis DKC2, the predominant Danish outbreak strain. Microbiol. Resour. Announc. 8, e01554–18 (2019).
Roetzer, A. et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLOS Med. 10, e1001387 (2013).
Bainomugisa, A. et al. A complete high-quality MinION nanopore assembly of an extensively drug-resistant Mycobacterium tuberculosis Beijing lineage strain identifies novel variation in repetitive PE/PPE gene regions. Microb. Genom. 4, 256719 (2018).
Iqbal, Z., Caccamo, M., Turner, I., Flicek, P. & McVean, G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat. Genet. 44, 226–232 (2012).
Yadon, A. N. et al. A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nat. Commun. 8, 588 (2017).
Yang, Y. et al. Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinformatics 34, 1666–1671 (2018).
Chen, M. L. et al. Deep learning predicts tuberculosis drug resistance status from genome sequencing data. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/275628v2 (2018).
Rajendran, V. & Sethumadhavan, R. Drug resistance mechanism of PncA in Mycobacterium tuberculosis. J. Biomol. Struct. Dyn. 32, 209–221 (2013).
Kavvas, E. S. et al. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat. Commun. 9, 4306 (2018).
Duchêne, S. et al. Genome-scale rates of evolutionary change in bacteria. Microb. Genom. 2, e000094 (2016).
Lee, R. S. et al. Reemergence and amplification of tuberculosis in the Canadian arctic. J. Infect. Dis. 211, 1905–1914 (2015).
Clark, T. G. et al. Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing. PLOS ONE 8, e83012 (2013).
Guthrie, J. L. et al. Genotyping and whole-genome sequencing to identify tuberculosis transmission to pediatric patients in British Columbia, Canada, 2005–2014. J. Infect. Dis. 218, 1155–1163 (2018).
Bryant, J. M. et al. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet. Respir. Med. 1, 786–792 (2013).
Guerra-Assunção, J. A. et al. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J. Infect. Dis. 211, 1154–1163 (2015).
Schürch, A. C. et al. The tempo and mode of molecular evolution of Mycobacterium tuberculosis at patient-to-patient scale. Infect. Genet. Evol. 10, 108–114 (2010).
Lieberman, T. D. et al. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat. Med. 22, 1470–1474 (2016).
Ford, C. B. et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 45, 784–790 (2013).
Ford, C. B. et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat. Genet. 43, 482–486 (2011).
Hatherell, H.-A. et al. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med. 14, 21 (2016). This is a systematic review of the potential for WGS in determining transmission of MTBC strains.
Verver, S. et al. Transmission of tuberculosis in a high incidence urban community in South Africa. Int. J. Epidemiol. 33, 351–357 (2004).
Bjorn-Mortensen, K. et al. Tracing Mycobacterium tuberculosis transmission by whole genome sequencing in a high incidence setting: a retrospective population-based study in East Greenland. Sci. Rep. 6, 33180 (2016).
Stimson, J. et al. Beyond the SNP threshold: identifying outbreak clusters using inferred transmissions. Mol. Biol. Evol. 36, 587–603 (2019).
Biek, R., Pybus, O. G., Lloyd-Smith, J. O. & Didelot, X. Measurably evolving pathogens in the genomic era. Trends Ecol. Evol. 30, 306–313 (2015).
Campbell, F. et al. outbreaker2: a modular platform for outbreak reconstruction. BMC Bioinformatics 19, 363 (2018).
Didelot, X., Gardy, J. & Colijn, C. Bayesian inference of infectious disease transmission from whole-genome sequence data. Mol. Biol. Evol. 31, 1869–1879 (2014).
Didelot, X., Fraser, C., Gardy, J. & Colijn, C. Genomic infectious disease epidemiology in partially sampled and ongoing outbreaks. Mol. Biol. Evol. 34, 997–1007 (2017).
De Maio, N., Worby, C. J., Wilson, D. J. & Stoesser, N. Bayesian reconstruction of transmission within outbreaks using genomic variants. PLOS Comput. Biol. 14, e1006117 (2018).
Klinkenberg, D., Backer, J. A., Didelot, X., Colijn, C. & Wallinga, J. Simultaneous inference of phylogenetic and transmission trees in infectious disease outbreaks. PLOS Comput. Biol. 13, e1005495 (2017).
Kühnert, D. et al. Tuberculosis outbreak investigation using phylodynamic analysis. Epidemics 25, 47–53 (2018).
Eldholm, V. et al. Armed conflict and population displacement as drivers of the evolution and dispersal of Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 113, 13881–13886 (2016).
Streicher, E. M. et al. Mycobacterium tuberculosis population structure determines the outcome of genetics-based second-line drug resistance testing. Antimicrob. Agents Chemother. 56, 2420–2427 (2012).
Folkvardsen, D. B. et al. Rifampin heteroresistance in Mycobacterium tuberculosis cultures as detected by phenotypic and genotypic drug susceptibility test methods. J. Clin. Microbiol. 51, 4220–4222 (2013).
Shamputa, I. C. et al. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir. Res. 7, 99 (2006).
Sobkowiak, B. et al. Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genomics 19, 613 (2018).
Gan, M., Liu, Q., Yang, C., Gao, Q. & Luo, T. Deep whole-genome sequencing to detect mixed infection of Mycobacterium tuberculosis. PLOS ONE 11, e0159029 (2016).
Votintseva, A. A. et al. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J. Clin. Microbiol. 55, 1285–1298 (2017).
Doyle, R. M. et al. Direct whole-genome sequencing of sputum accurately identifies drug-resistant Mycobacterium tuberculosis faster than MGIT culture sequencing. J. Clin. Microbiol. 56, e00666–18 (2018).
Doughty, E. L., Sergeant, M. J., Adetifa, I., Antonio, M. & Pallen, M. J. Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer. PeerJ 2, e585 (2014).
Phelan, J. E. et al. Recombination in pe/ppe genes contributes to genetic variation in Mycobacterium tuberculosis lineages. BMC Genomics 17, 151 (2016).
Reisner, B. S., Gatson, A. M. & Woods, G. L. Evaluation of mycobacteria growth indicator tubes for susceptibility testing of Mycobacterium tuberculosis to isoniazid and rifampin. Diagn. Microbiol. Infect. Dis. 22, 325–329 (1995).
Strydom, K. et al. Comparison of three commercial molecular assays for detection of rifampin and isoniazid resistance among Mycobacterium tuberculosis isolates in a high-HIV-prevalence setting. J. Clin. Microbiol. 53, 3032–3034 (2015).
Nathavitharana, R. R. et al. Multicenter noninferiority evaluation of Hain GenoType MTBDRplus version 2 and Nipro NTM+MDRTB line probe assays for detection of rifampin and isoniazid resistance. J. Clin. Microbiol. 54, 1624–1630 (2016).
Mitarai, S. et al. Comprehensive multicenter evaluation of a new line probe assay kit for identification of Mycobacterium species and detection of drug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 50, 884–890 (2012).
Hillemann, D., Rüsch-Gerdes, S. & Richter, E. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 47, 1767–1772 (2009).
Tagliani, E. et al. Diagnostic performance of the new version (v2.0) of GenoType MTBDR sl assay for detection of resistance to fluoroquinolones and second-line injectable drugs: a multicenter study. J. Clin. Microbiol. 53, 2961–2969 (2015).
Ng, K. C. et al. Potential application of digitally linked tuberculosis diagnostics for real-time surveillance of drug-resistant tuberculosis transmission: validation and analysis of test results. JMIR Med. Inform. 6, e12 (2018).
Chakravorty, S. et al. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. mBio 8, e00812–17 (2017).
Ng, K. C. S. et al. Xpert Ultra can unambiguously identify specific rifampin resistance-conferring mutations. J. Clin. Microbiol. 56, e00686–18 (2018).
Molina-Moya, B. et al. Diagnostic accuracy study of multiplex PCR for detecting tuberculosis drug resistance. J. Infect. 71, 220–230 (2015).
Hillemann, D., Haasis, C., Andres, S., Behn, T. & Kranzer, K. Validation of the FluoroType MTBDR assay for detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 56, e00072–18 (2018).
Pang, Y. et al. Rapid diagnosis of MDR and XDR tuberculosis with the MeltPro TB assay in China. Sci. Rep. 6, 25330 (2016).
Kaswa, M. K. et al. Pseudo-outbreak of pre-extensively drug-resistant (Pre-XDR) tuberculosis in Kinshasa: collateral damage caused by false detection of fluoroquinolone resistance by GenoType MTBDRsl. J. Clin. Microbiol. 52, 2876–2880 (2014).
Ajileye, A. et al. Some synonymous and nonsynonymous gyrA mutations in Mycobacterium tuberculosis lead to systematic false-positive fluoroquinolone resistance results with the Hain GenoType MTBDRsl assays. Antimicrob. Agents Chemother. 61, e02169–16 (2017).
Colman, R. E. et al. Detection of low-level mixed-population drug resistance in Mycobacterium tuberculosis using high fidelity amplicon sequencing. PLOS ONE 10, e0126626 (2015).
Colman, R. E. et al. Rapid drug susceptibility testing of drug-resistant Mycobacterium tuberculosis isolates directly from clinical samples by use of amplicon sequencing: a proof-of-concept study. J. Clin. Microbiol. 54, 2058–2067 (2016).
Makhado, N. A. et al. Outbreak of multidrug-resistant tuberculosis in South Africa undetected by WHO-endorsed commercial tests: an observational study. Lancet Infect. Dis. 18, 1350–1359 (2018).
Tagliani, E. et al. Culture and next-generation sequencing-based drug susceptibility testing unveil high levels of drug-resistant-TB in Djibouti: results from the first national survey. Sci. Rep. 7, 17672 (2017).
Gagneux, S. Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 202–213 (2018).
Acknowledgements
C.J.M. and L.R. are also affiliated with BCCM/ITM Mycobacterial Culture Collection, Institute of Tropical Medicine, Antwerp, Belgium. J.G. is also affiliated with the BC Centre for Disease Control, Vancouver, Canada. B.O.-A. is also affiliated with the Center for Global Health Security and Diplomacy, Ottawa, Canada. M.S. is also affiliated with the University of Arizona, Tucson, AZ, USA. I.C. is also affiliated with the CIBER in Epidemiology and Public Health, Spain. C.J.M., B.O.-A., L.R. and B.C.d.J. are supported by a European Research Council grant (INTERRUPTB; no. 311725). I.C. and G.A.G. are supported by a European Research Council grant (TB-ACCELERATE; no. 638553). T.C.R. receives salary support from the not-for-profit organization Foundation for Innovative New Diagnostics (the terms of this arrangement have been reviewed and approved by the University of California, San Diego). T.M.W. is an NIHR Academic Clinical Lecturer. J.L.G. and J.G. receive funding from the University of British Columbia, Vancouver, Canada. T.A.K., C.U., V.D. and S.N. receive funding from the German Center for Infection Research (DZIF) and are funded by the Deutsche Forschungsgemeinschaft (German Research Foundation) under Germany’s Excellence Strategy (EXC 22167–390884018). L.V., T.H.H. and A.V.R. are funded by FWO Odysseus G0F8316N. M.R.F. is supported by the US National Institutes of Health BD2K K01 (MRF ES026835). P.S. is supported by the Agence Nationale de la Recherche (ANR-16-CE35-0009).
Reviewer information
Nature Reviews Microbiology thanks T. McHugh, V. Sintchenko, and other anonymous reviewer(s), for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
C.J.M., G.A.G., T.A.K., L.V., A.D., M.E., M.R.F., J.L.G., K.L., P.M., B.O.-A., V.D., P.S., A.S., C.U., D.v.S., Y.Z., M.S., J.G., D.M.C., S.N., I.C. and A.V.R. researched the data for the article. C.J.M., G.A.G., T.A.K., L.V., A.D., M.E., M.R.F., J.L.G., K.L., P.M., B.O.-A., V.D., P.S., A.S., C.U., D.v.S. Y.Z., M.d.V., S.G., T.H.H., L.R., E.T., T.M.W., R.M.W., M.S., J.G., D.M.C., S.N., I.C. and A.V.R. substantially contributed to the discussion of the content. C.J.M., G.A.G., T.A.K., L.V., A.D., M.E., M.R.F., J.L.G., K.L., P.M., B.O.-A., V.D., P.S., A.S., C.U., D.v.S., Y.Z., M.S., J.G., D.M.C., S.N., I.C. and A.V.R. wrote the article. All authors reviewed and edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
P.S. was a consultant for Genoscreen. All other authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Human, Heredity and Health in Africa Consortium: https://h3abionet.org
ReSeqTB: http://www.reseqtb.org
TORCH consortium: https://torch-consortium.com/vliruos
Supplementary information
Glossary
- Mycobacterium tuberculosis complex
-
(MTBC). The genetically related group of organisms within the genus Mycobacterium that cause tuberculosis in humans or animals.
- Drug susceptibility testing
-
(DST). A procedure to determine if clinical isolates are resistant to antibiotics either by testing the inhibition in culture (phenotypic DST) or by identifying drug resistance-associated mutations (genotypic DST).
- Source investigation
-
The first case in a group of related individuals that transmitted the disease. Usually, identified during the development of an epidemiological investigation.
- Löwenstein–Jensen
-
A selective culture solid medium commonly used to isolate Mycobacterium tuberculosis complex strains.
- Mycobacteria Growth Indicator Tube
-
A tube that contains mycobacteria-selective culture liquid medium and is usually coupled to an automated instrument to read the results.
- PE and PPE gene families
-
Families of genes that encode virulence factors in Mycobacterium tuberculosis complex strains. They have signature (proline)–proline–glutamate ((P)PE) motifs at their amino terminus.
- Core genome MLST
-
A scheme that converts genome-wide SNP data into an allele-numbering system using a preselected set of core genes.
- Whole genome MLST
-
A scheme that converts genome-wide SNP data into an allele-numbering system using a preselected set of core genes and additional accessory genes.
- Contact tracing
-
The identification of possible contacts that interacted with an infected person (index case), often through questionnaires and interviews.
- Mycobacterial interspersed repetitive unit variable-number tandem repeat
-
Mycobacterium tuberculosis complex (MTBC)-specific variable tandem repeat locus used to genotype MTBC strains.
- WGS pipelines
-
The bioinformatics section of the whole genome sequencing workflow, starting from raw sequencing files through to SNP calling and analyses.
- WGS workflows
-
All steps involved (from culturing to SNP calling and analyses) for whole genome sequencing of an isolate.
- BioCompute Object
-
(BCO). A framework for standardized reporting of computational parameters for a whole genome sequencing pipeline.
- Spoligotyping
-
A PCR-based approach based on the amplification of spacers in the CRISPR region of the Mycobacterium tuberculosis complex (MTBC). It is used for genotyping MTBC strains.
- Effective reproduction number
-
The average number of secondary cases per infectious case.
- Microevolution
-
Genetic changes within a population, resulting in separate subpopulations.
Rights and permissions
About this article
Cite this article
Meehan, C.J., Goig, G.A., Kohl, T.A. et al. Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol 17, 533–545 (2019). https://doi.org/10.1038/s41579-019-0214-5
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-019-0214-5
This article is cited by
-
On the onset and dispersal of a major MDR TB clone among HIV-negative patients, Tunisia
Antimicrobial Resistance & Infection Control (2024)
-
What is New in the Diagnosis of Childhood Tuberculosis?
Indian Journal of Pediatrics (2024)
-
Performance evaluation of core genome multilocus sequence typing for genotyping of Mycobacterium tuberculosis strains in China: based on multicenter, population-based collection
European Journal of Clinical Microbiology & Infectious Diseases (2024)
-
Characterization of microbial communities in urban subway: connotation for indoor environment quality and public health
Air Quality, Atmosphere & Health (2024)
-
Insidious transmission of Mycobacterium tuberculosis in Ordos, China: a molecular epidemiology study
European Journal of Clinical Microbiology & Infectious Diseases (2024)