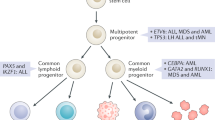
Abstract
Somatic mutations occur spontaneously in normal individuals and accumulate throughout life. These genetic modifications contribute to progressive ageing phenotypes and are directly involved in cancer development. However, a growing number of studies of Mendelian haematopoietic disorders indicate that somatic genetic events can offset the pathogenic effect of germline mutations at the cellular level, leading to genetic mosaicism and, in some cases, resulting in a milder disease phenotype. Notably, spontaneous genetic events that confer a positive effect on cells do not always benefit the individual, for whom the effects can be neutral or even clinically detrimental. These somatic genetic rescue events have important diagnostic, therapeutic and clinical consequences and constitute valuable models for studying the differentiation and/or homeostasis of haematopoietic lineages.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lee-Six, H. et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Jacobs, K. B. et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat. Genet. 44, 651–658 (2012).
Laurie, C. C. et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 44, 642–650 (2012).
Osorio, F. G. et al. Somatic mutations reveal lineage relationships and age-related mutagenesis in human hematopoiesis. Cell Rep. 25, 2308–2316 (2018). The studies by Lee-Six et al. and Osorio et al. use ultra-deep sequencing to quantify the rate of somatic mutations accumulating over time in HSCs and circulating blood cells from normal individuals and provide insight into the differentiation pathways of the haematopoietic lineages.
Mufti, G. J. & Marsh, J. C. W. Somatic mutations in aplastic anemia. Hematol. Oncol. Clin. North Am. 32, 595–607 (2018).
Jaiswal, S. et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017).
Welch, J. S. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012).
Youssoufian, H. & Pyeritz, R. E. Mechanisms and consequences of somatic mosaicism in humans. Nat. Rev. Genet. 3, 748–758 (2002).
Campbell, I. M., Shaw, C. A., Stankiewicz, P. & Lupski, J. R. Somatic mosaicism: implications for disease and transmission genetics. Trends Genet. 31, 382–392 (2015).
Wegman-Ostrosky, T. & Savage, S. A. The genomics of inherited bone marrow failure: from mechanism to the clinic. Br. J. Haematol. 177, 526–542 (2017).
Picard, C. et al. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J. Clin. Immunol. 38, 96–128 (2018).
Donadieu, J., Beaupain, B., Fenneteau, O. & Bellanne-Chantelot, C. Congenital neutropenia in the era of genomics: classification, diagnosis, and natural history. Br. J. Haematol. 179, 557–574 (2017).
Forterre, P. Darwin’s goldmine is still open: variation and selection run the world. Front. Cell. Infect. Microbiol. 2, 106 (2012).
Hirschhorn, R., Yang, D. R., Israni, A., Huie, M. L. & Ownby, D. R. Somatic mosaicism for a newly identified splice-site mutation in a patient with adenosine deaminase-deficient immunodeficiency and spontaneous clinical recovery. Am. J. Hum. Genet. 55, 59–68 (1994). This study is among the first to report SGR leading to a clear-cut cell and clinical correction of an inherited haematopoietic disease.
Hirschhorn, R. et al. Spontaneous in vivo reversion to normal of an inherited mutation in a patient with adenosine deaminase deficiency. Nat. Genet. 13, 290–295 (1996).
Stephan, V. et al. Atypical X-linked severe combined immunodeficiency due to possible spontaneous reversion of the genetic defect in T cells. N. Engl. J. Med. 335, 1563–1567 (1996).
Ellis, N. A. et al. Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am. J. Hum. Genet. 57, 1019–1027 (1995).
Foucault, F. et al. Characterization of a new BLM mutation associated with a topoisomerase II alpha defect in a patient with Bloom’s syndrome. Hum. Mol. Genet. 6, 1427–1434 (1997).
Waisfisz, Q. et al. Spontaneous functional correction of homozygous Fanconi anaemia alleles reveals novel mechanistic basis for reverse mosaicism. Nat. Genet. 22, 379–383 (1999).
Lo Ten Foe, J. R. et al. Somatic mosaicism in Fanconi anemia: molecular basis and clinical significance. Eur. J. Hum. Genet. 5, 137–148 (1997).
Suzuki, S. et al. Somatic recombination underlies frequent revertant mosaicism in Loricrin keratoderma. Life Sci. Alliance 2, e201800284 (2019).
Lim, Y. H., Fisher, J. M. & Choate, K. A. Revertant mosaicism in genodermatoses. Cell. Mol. Life Sci. 74, 2229–2238 (2017).
Burrow, K. L. et al. Dystrophin expression and somatic reversion in prednisone-treated and untreated Duchenne dystrophy. CIDD Study Group. Neurology 41, 661–666 (1991).
Klein, C. J. et al. Somatic reversion/suppression in Duchenne muscular dystrophy (DMD): evidence supporting a frame-restoring mechanism in rare dystrophin-positive fibers. Am. J. Hum. Genet. 50, 950–959 (1992).
Fanin, M. et al. Dystrophin-positive fibers in Duchenne dystrophy: origin and correlation to clinical course. Muscle Nerve 18, 1115–1120 (1995).
Punetha, J. et al. Somatic mosaicism due to a reversion variant causing hemi-atrophy: a novel variant of dystrophinopathy. Eur. J. Hum. Genet. 24, 1511–1514 (2016).
Poudrier, J., Lettre, F., Scriver, C. R., Larochelle, J. & Tanguay, R. M. Different clinical forms of hereditary tyrosinemia (type I) in patients with identical genotypes. Mol. Genet. Metab. 64, 119–125 (1998).
Dreumont, N. et al. A missense mutation (Q279R) in the fumarylacetoacetate hydrolase gene, responsible for hereditary tyrosinemia, acts as a splicing mutation. BMC Genet. 2, 9 (2001).
Demers, S. I., Russo, P., Lettre, F. & Tanguay, R. M. Frequent mutation reversion inversely correlates with clinical severity in a genetic liver disease, hereditary tyrosinemia. Hum. Pathol. 34, 1313–1320 (2003).
Federico, C. et al. Somatic mosaicism with reversion to normality of a mutated transthyretin allele related to a familial amyloidotic polyneuropathy. Hum. Genet. 136, 867–873 (2017).
Scalet, D. et al. The somatic FAH C.1061C>A change counteracts the frequent FAH c.1062 + 5G>A mutation and permits U1snRNA-based splicing correction. J. Hum. Genet. 63, 683–686 (2018).
Ikeda, H. et al. Genetic reversion in an acute myelogenous leukemia cell line from a Fanconi anemia patient with biallelic mutations in BRCA2. Cancer Res. 63, 2688–2694 (2003).
Gross, M. et al. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet. Genome Res. 98, 126–135 (2002).
George, J. W. et al. Restoration of nucleotide excision repair in a helicase-deficient XPD mutant from intragenic suppression by a trichothiodystrophy mutation. Mol. Cell. Biol. 21, 7355–7365 (2001).
Bogliolo, M. et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 92, 800–806 (2013).
Edwards, S. L. et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451, 1111–1115 (2008).
Kondrashova, O. et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 7, 984–998 (2017).
Quigley, D. et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 7, 999–1005 (2017).
Dulskas, A. et al. A case of gastric cancer metastasis to the breast in a female with BRCA2 germline mutation and literature review. Acta Chir. Belg. 119, 59–63 (2019).
Bouwman, P. & Jonkers, J. Molecular pathways: how can BRCA-mutated tumors become resistant to PARP inhibitors? Clin. Cancer Res. 20, 540–547 (2014).
Gillies, R. J., Verduzco, D. & Gatenby, R. A. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat. Rev. Cancer 12, 487–493 (2012).
Patch, A. M. et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015).
Moncada-Velez, M. et al. Somatic mosaicism caused by monoallelic reversion of a mutation in T cells of a patient with ADA-SCID and the effects of enzyme replacement therapy on the revertant phenotype. Scand. J. Immunol. 74, 471–481 (2011).
Liu, P. et al. Immunologic reconstitution during PEG-ADA therapy in an unusual mosaic ADA deficient patient. Clin. Immunol. 130, 162–174 (2009).
Ariga, T. et al. T cell lines from 2 patients with adenosine deaminase (ADA) deficiency showed the restoration of ADA activity resulted from the reversion of an inherited mutation. Blood 97, 2896–2899 (2001).
Crestani, E. et al. RAG1 reversion mosaicism in a patient with Omenn syndrome. J. Clin. Immunol. 34, 551–554 (2014).
Brigida, I. et al. T cell defects in patients with ARPC1B germline mutations account for their combined immunodeficiency. Blood 132, 2362–2374 (2018).
Ellis, N. A., Ciocci, S. & German, J. Back mutation can produce phenotype reversion in Bloom syndrome somatic cells. Hum. Genet. 108, 167–173 (2001).
Marin, A. V. et al. Primary T cell immunodeficiency with functional revertant somatic mosaicism in CD247. J. Allergy Clin. Immunol. 139, 347–349 (2017).
Stray-Pedersen, A. et al. Primary immunodeficiency diseases: genomic approaches delineate heterogeneous Mendelian disorders. J. Allergy Clin. Immunol. 139, 232–245 (2017).
Perdigones, N. et al. Clonal hematopoiesis in patients with dyskeratosis congenita. Am. J. Hematol. 91, 1227–1233 (2016).
Jing, H. et al. Somatic reversion in dedicator of cytokinesis 8 immunodeficiency modulates disease phenotype. J. Allergy Clin. Immunol. 133, 1667–1675 (2014).
Kienzler, A. K. et al. Hypomorphic function and somatic reversion of DOCK8 cause combined immunodeficiency without hyper-IgE. Clin. Immunol. 163, 17–21 (2016).
Kuijpers, T. W. et al. A reversion of an IL2RG mutation in combined immunodeficiency providing competitive advantage to the majority of CD8+ T cells. Haematologica 98, 1030–1038 (2013).
Hsu, A. P. et al. IL2RG reversion event in a common lymphoid progenitor leads to delayed diagnosis and milder phenotype. J. Clin. Immunol. 35, 449–453 (2015).
Speckmann, C. et al. Clinical and immunologic consequences of a somatic reversion in a patient with X-linked severe combined immunodeficiency. Blood 112, 4090–4097 (2008).
Kawai, T. et al. Multiple reversions of an IL2RG mutation restore T cell function in an X-linked severe combined immunodeficiency patient. J. Clin. Immunol. 32, 690–697 (2012).
Tone, Y. et al. Somatic revertant mosaicism in a patient with leukocyte adhesion deficiency type 1. Blood 109, 1182–1184 (2007).
Uzel, G. et al. Reversion mutations in patients with leukocyte adhesion deficiency type-1 (LAD-1). Blood 111, 209–218 (2008).
Le Guen, T. et al. An in vivo genetic reversion highlights the crucial role of Myb-Like, SWIRM, and MPN domains 1 (MYSM1) in human hematopoiesis and lymphocyte differentiation. J. Allergy Clin. Immunol. 136, 1619–1626 (2015). This report describes the occurrence of SGR in MYSM1 deficiency that leads to a full correction of a disease phenotype at both the cellular and clinical levels.
Palendira, U. et al. Expansion of somatically reverted memory CD8+ T cells in patients with X-linked lymphoproliferative disease caused by selective pressure from Epstein–Barr virus. J. Exp. Med. 209, 913–924 (2012).
Ariga, T. et al. Spontaneous in vivo reversion of an inherited mutation in the Wiskott–Aldrich syndrome. J. Immunol. 166, 5245–5249 (2001).
Boztug, K. et al. Large granular lymphocyte proliferation and revertant mosaicism: two rare events in a Wiskott–Aldrich syndrome patient. Haematologica 92, e43–e45 (2007).
Lutskiy, M. I., Beardsley, D. S., Rosen, F. S. & Remold-O’Donnell, E. Mosaicism of NK cells in a patient with Wiskott–Aldrich syndrome. Blood 106, 2815–2817 (2005).
Stewart, D. M., Candotti, F. & Nelson, D. L. The phenomenon of spontaneous genetic reversions in the Wiskott–Aldrich syndrome: a report of the workshop of the ESID Genetics Working Party at the XIIth Meeting of the European Society for Immunodeficiencies (ESID). Budapest, Hungary October 4–7, 2006. J. Clin. Immunol. 27, 634–639 (2007).
Xie, J. W. et al. In vivo reversion of an inherited mutation in a Chinese patient with Wiskott–Aldrich syndrome. Hum. Immunol. 76, 406–413 (2015).
Kalb, R. et al. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am. J. Hum. Genet. 80, 895–910 (2007).
Davis, B. R. et al. Unprecedented diversity of genotypic revertants in lymphocytes of a patient with Wiskott–Aldrich syndrome. Blood 111, 5064–5067 (2008). These authors report up to 34 different SGR events in a single patient with Wiskott–Aldrich syndrome, demonstrating that distinct somatic mutations can accumulate in different clones from the same individual.
Bluteau, O. et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood 131, 717–732 (2018).
Buonocore, F. et al. Somatic mutations and progressive monosomy modify SAMD9-related phenotypes in humans. J. Clin. Invest. 127, 1700–1713 (2017). This study reports patients with MIRAGE syndrome, caused by germline heterozygous gof mutations in SAMD9, in whom somatic monosomy 7, interstitial 7q deletion or lof mutation in SAMD9 in blood cells represents recurrent SGR that eliminates the deleterious effect of the germline mutation and leads to clinical improvement.
Shima, H. et al. Two patients with MIRAGE syndrome lacking haematological features: role of somatic second-site reversion SAMD9 mutations. J. Med. Genet. 55, 81–85 (2018).
Schwartz, J. R. et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat. Commun. 8, 1557 (2017).
Schwartz, J. R. et al. Germline SAMD9 mutation in siblings with monosomy 7 and myelodysplastic syndrome. Leukemia 31, 1827–1830 (2017).
Narumi, S. et al. SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat. Genet. 48, 792–797 (2016).
Pastor, V. B. et al. Constitutional SAMD9L mutations cause familial myelodysplastic syndrome and transient monosomy 7. Haematologica 103, 427–437 (2018).
Wong, J. C. et al. Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight 3, 121086 (2018).
Tesi, B. et al. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood 129, 2266–2279 (2017).
Chen, D. H. et al. Ataxia-pancytopenia syndrome is caused by missense mutations in SAMD9L. Am. J. Hum. Genet. 98, 1146–1158 (2016).
Wada, T. et al. Detection of T lymphocytes with a second-site mutation in skin lesions of atypical X-linked severe combined immunodeficiency mimicking Omenn syndrome. Blood 112, 1872–1875 (2008).
Blackburn, E. H., Greider, C. W. & Szostak, J. W. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12, 1133–1138 (2006).
Maryoung, L. et al. Somatic mutations in telomerase promoter counterbalance germline loss-of-function mutations. J. Clin. Invest. 127, 982–986 (2017).
Gutierrez-Rodrigues, F. et al. Pathogenic TERT promoter variants in telomere diseases. Genet. Med. https://doi.org/10.1038/s41436-018-0385-x (2018). Maryoung et al. and Guiterrez-Rodrigues et al. describe what are thought to be the first cases of indirect SGR, in which somatic mutations that activate the TERT promoter in blood cells counteract a germline lof mutation in a gene implicated in telomere maintenance (TERC and PARN).
Heidenreich, B. & Kumar, R. TERT promoter mutations in telomere biology. Mutat. Res. 771, 15–31 (2017).
Horn, S. et al. TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013).
Chiba, K. et al. Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife 4, e07918 (2015).
Wada, T. et al. Somatic mosaicism in Wiskott–Aldrich syndrome suggests in vivo reversion by a DNA slippage mechanism. Proc. Natl Acad. Sci. USA 98, 8697–8702 (2001).
Pinto, F. O. et al. Diagnosis of Fanconi anemia in patients with bone marrow failure. Haematologica 94, 487–495 (2009).
Gregory, J. J. Jr. et al. Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc. Natl Acad. Sci. USA 98, 2532–2537 (2001).
Konno, A. et al. Differential contribution of Wiskott–Aldrich syndrome protein to selective advantage in T- and B-cell lineages. Blood 103, 676–678 (2004).
Alder, J. K. et al. Exome sequencing identifies mutant TINF2 in a family with pulmonary fibrosis. Chest 147, 1361–1368 (2015).
Asur, R. S. et al. Somatic mosaicism of an intragenic FANCB duplication in both fibroblast and peripheral blood cells observed in a Fanconi anemia patient leads to milder phenotype. Mol. Genet. Genom. Med. 6, 77–91 (2018).
Virts, E. L. et al. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum. Mol. Genet. 24, 5093–5108 (2015).
Nishikomori, R. et al. X-Linked ectodermal dysplasia and immunodeficiency caused by reversion mosaicism of NEMO reveals a critical role for NEMO in human T cell development and/or survival. Blood 103, 4565–4572 (2004).
Jiang, J. et al. Molecular and immunological characterization of DNA ligase IV deficiency. Clin. Immunol. 163, 75–83 (2016).
Lapunzina, P. & Monk, D. The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer. Biol. Cell 103, 303–317 (2011).
Jongmans, M. C. et al. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am. J. Hum. Genet. 90, 426–433 (2012). These authors report, thought to be for the first time in telomeropathy, the somatic reversion of the TERC mutated allele to wild type through mitotic recombination events in blood cells from four individuals with germline TERC mutations.
Jongmans, M. C. J. et al. Somatic reversion events point towards RPL4 as a novel disease gene in a condition resembling Diamond-Blackfan anemia. Haematologica 103, e607–e609 (2018).
Venugopal, P. et al. Self-reverting mutations partially correct the blood phenotype in a Diamond Blackfan anemia patient. Haematologica 102, e506–e509 (2017).
Ly, P. & Cleveland, D. W. Rebuilding chromosomes after catastrophe: emerging mechanisms of chromothripsis. Trends Cell Biol. 27, 917–930 (2017).
McDermott, D. H. et al. Chromothriptic cure of WHIM syndrome. Cell 160, 686–699 (2015). The authors report that even dramatic modification of the genome through chromothripsis can restore haematopoiesis by eliminating a dominant deleterious mutation in CXCR4.
Vulliamy, T. et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl Acad. Sci. USA 105, 8073–8078 (2008).
Boocock, G. R. et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 33, 97–101 (2003).
Finch, A. J. et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 25, 917–929 (2011).
Weis, F. et al. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 22, 914–919 (2015).
Warren, A. J. Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv. Biol. Regul. 67, 109–127 (2018).
Maserati, E. et al. Shwachman syndrome as mutator phenotype responsible for myeloid dysplasia/neoplasia through karyotype instability and chromosomes 7 and 20 anomalies. Genes, chromosomes and cancer 45, 375–382 (2006).
Nacci, L. et al. Parental origin of the deletion del(20q) in Shwachman-Diamond patients and loss of the paternally derived allele of the imprinted L3MBTL1 gene. Genes Chromosomes Cancer 56, 51–58 (2017).
Pressato, B. et al. Deletion of chromosome 20 in bone marrow of patients with Shwachman-Diamond syndrome, loss of the EIF6 gene and benign prognosis. Br. J. Haematol. 157, 503–505 (2012).
Valli, R. et al. Different loss of material in recurrent chromosome 20 interstitial deletions in Shwachman-Diamond syndrome and in myeloid neoplasms. Mol. Cytogenet. 6, 56 (2013).
Menne, T. F. et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 39, 486–495 (2007).
Ceccaldi, R. et al. Spontaneous abrogation of the G(2)DNA damage checkpoint has clinical benefits but promotes leukemogenesis in Fanconi anemia patients. J. Clin. Invest. 121, 184–194 (2011).
Biesecker, L. G. & Spinner, N. B. A genomic view of mosaicism and human disease. Nat. Rev. Genet. 14, 307–320 (2013).
Hofer, T. & Rodewald, H. R. Differentiation-based model of hematopoietic stem cell functions and lineage pathways. Blood 132, 1106–1113 (2018).
Shpall, E. J., Champlin, R. & Glaspy, J. A. Effect of CD34+ peripheral blood progenitor cell dose on hematopoietic recovery. Biol. Blood Marrow Transplant. 4, 84–92 (1998).
Wada, T. et al. Second-site mutation in the Wiskott–Aldrich syndrome (WAS) protein gene causes somatic mosaicism in two WAS siblings. J. Clin. Invest. 111, 1389–1397 (2003).
Ban, S. A. et al. Combined immunodeficiency evolving into predominant CD4+ lymphopenia caused by somatic chimerism in JAK3. J. Clin. Immunol. 34, 941–953 (2014).
Hoshino, A. et al. Modification of cellular and humoral immunity by somatically reverted T cells in X-linked lymphoproliferative syndrome type 1. J. Allergy Clin. Immunol. 143, 421–424 (2019).
Paul, S., Million-Weaver, S., Chattopadhyay, S., Sokurenko, E. & Merrikh, H. Accelerated gene evolution through replication-transcription conflicts. Nature 495, 512–515 (2013).
Sankar, T. S., Wastuwidyaningtyas, B. D., Dong, Y., Lewis, S. A. & Wang, J. D. The nature of mutations induced by replication-transcription collisions. Nature 535, 178–181 (2016).
Gangloff, S. & Arcangioli, B. DNA repair and mutations during quiescence in yeast. FEMS Yeast Res. 17, fox002 (2017).
Hastings, P. J., Lupski, J. R., Rosenberg, S. M. & Ira, G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 10, 551–564 (2009).
Blazquez-Moreno, A. et al. Analysis of the recovery of CD247 expression in a PID patient: insights into the spontaneous repair of defective genes. Blood 130, 1205–1208 (2017).
Orkin, S. H. & Zon, L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008).
Zuniga-Pflucker, J. C. T cell development made simple. Nat. Rev. Immunol. 4, 67–72 (2004).
Klein, L., Kyewski, B., Allen, P. M. & Hogquist, K. A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14, 377–391 (2014).
Okhrimenko, A. et al. Human memory T cells from the bone marrow are resting and maintain long-lasting systemic memory. Proc. Natl Acad. Sci. USA 111, 9229–9234 (2014).
Bousso, P. et al. Diversity, functionality, and stability of the T cell repertoire derived in vivo from a single human T cell precursor. Proc. Natl Acad. Sci. USA 97, 274–278 (2000).
Notarangelo, L. D., Kim, M. S., Walter, J. E. & Lee, Y. N. Human RAG mutations: biochemistry and clinical implications. Nat. Rev. Immunol. 16, 234–246 (2016).
Rawlings, D. J. et al. Mutation of unique region of Bruton’s tyrosine kinase in immunodeficient XID mice. Science 261, 358–361 (1993).
Tsukada, S. et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell 72, 279–290 (1993).
Okano, T. et al. Maternal T and B cell engraftment in two cases of X-linked severe combined immunodeficiency with IgG1 gammopathy. Clin. Immunol. 183, 112–120 (2017).
Kobrynski, L. J. & Abramowsky, C. Monoclonal IgA gammopathy due to maternal B cells in an infant with severe combined immunodeficiency (SCID) prior to hematopoietic stem cell transplantation. J. Pediatr. Hematol. Oncol. 28, 53–56 (2006).
Morinishi, Y. et al. Identification of severe combined immunodeficiency by T cell receptor excision circles quantification using neonatal guthrie cards. J. Pediatr. 155, 829–833 (2009).
Lanfranchi, A. et al. Maternal T cell engraftment impedes with diagnosis of a SCID-ADA patient. Clin. Immunol. 193, 118–120 (2018).
Wahlstrom, J. et al. Transplacental maternal engraftment and posttransplantation graft-versus-host disease in children with severe combined immunodeficiency. J. Allergy Clin. Immunol. 139, 628–633 (2017).
Lev, A. et al. Co-existence of clonal expanded autologous and transplacental-acquired maternal T cells in recombination activating gene-deficient severe combined immunodeficiency. Clin. Exp. Immunol. 176, 380–386 (2014).
Cattaneo, F. et al. Hypomorphic Janus kinase 3 mutations result in a spectrum of immune defects, including partial maternal T cell engraftment. J. Allergy Clin. Immunol. 131, 1136–1145 (2013).
Dvorak, C. C. et al. Maternal T cell engraftment associated with severe hemophagocytosis of the bone marrow in untreated X-linked severe combined immunodeficiency. J. Pediatr. Hematol. Oncol. 30, 396–400 (2008).
Kellermayer, R. et al. A novel IL2RG mutation associated with maternal T lymphocyte engraftment in a patient with severe combined immunodeficiency. J. Hum. Genet. 51, 495–497 (2006).
Muller, S. M. et al. Transplacentally acquired maternal T lymphocytes in severe combined immunodeficiency: a study of 121 patients. Blood 98, 1847–1851 (2001).
Villa, A. et al. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood 97, 81–88 (2001).
Sottini, A. et al. Engrafted maternal T cells in a severe combined immunodeficiency patient express T cell receptor variable beta segments characterized by a restricted V-D-J junctional diversity. Blood 85, 2105–2113 (1995).
Fischer, A., Notarangelo, L. D., Neven, B., Cavazzana, M. & Puck, J. M. Severe combined immunodeficiencies and related disorders. Nat. Rev. Dis. Primers 1, 15061 (2015).
Panchal, N., Booth, C., Cannons, J. L. & Schwartzberg, P. L. X-linked lymphoproliferative disease type 1: a clinical and molecular perspective. Front. Immunol. 9, 666 (2018).
May, M. Mutations to the rescue. Nat. Med. 17, 405–407 (2011).
Cavazzana-Calvo, M. et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288, 669–672 (2000). This study reports successful gene therapy of SCID X1 based on the selective advantage provided by wild-type gene integration, which mimics natural revertants.
Aiuti, A. et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science 296, 2410–2413 (2002). This study reports successful gene therapy of ADA SCID, based on the selective advantage provided by wild-type gene integration, which mimics natural revertants.
Hacein-Bey-Abina, S. et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 363, 355–364 (2010).
Clarke, E. L. et al. T cell dynamics and response of the microbiota after gene therapy to treat X-linked severe combined immunodeficiency. Genome Med. 10, 70 (2018).
Davidsson, J. et al. SAMD9 and SAMD9L in inherited predisposition to ataxia, pancytopenia, and myeloid malignancies. Leukemia 32, 1106–1115 (2018).
Inaba, T., Honda, H. & Matsui, H. The enigma of monosomy 7. Blood 131, 2891–2898 (2018).
Nagamachi, A. et al. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell 24, 305–317 (2013).
Nagata, Y. et al. Germline loss of function SAMD9 and SAMD9L alterations in adult myelodysplastic syndromes. Blood 132, 2309–2313 (2018).
Andres-Lencina, J. J. et al. TERT promoter mutation subtypes and survival in stage I and II melanoma patients. Int. J. Cancer 144, 1027–1036 (2019).
Bojesen, S. E. et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 45, 371–384 (2013).
Heidenreich, B. et al. Telomerase reverse transcriptase promoter mutations in primary cutaneous melanoma. Nat. Commun. 5, 3401 (2014).
Heidenreich, B., Rachakonda, P. S., Hemminki, K. & Kumar, R. TERT promoter mutations in cancer development. Curr. Opin. Genet. Dev. 24, 30–37 (2014).
Heidenreich, B. et al. TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget 6, 10617–10633 (2015).
Hosen, I. et al. Mutations in TERT promoter and FGFR3 and telomere length in bladder cancer. Int. J. Cancer 137, 1621–1629 (2015).
Hosen, I. et al. TERT promoter mutations in clear cell renal cell carcinoma. Int. J. Cancer 136, 2448–2452 (2015).
Huang, D. S. et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur. J. Cancer 51, 969–976 (2015).
Nagore, E. et al. TERT promoter mutations in melanoma survival. Int. J. Cancer 139, 75–84 (2016).
Rachakonda, P. S. et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc. Natl Acad. Sci. USA 110, 17426–17431 (2013).
Rachakonda, S. et al. Telomere length, telomerase reverse transcriptase promoter mutations, and melanoma risk. Genes Chromosomes Cancer 57, 564–572 (2018).
Simon, M. et al. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro-oncology 17, 45–52 (2015).
Aiuti, A. et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science 341, 1233151 (2013).
Min, Y. L. et al. CRISPR–Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 5, eaav4324 (2019).
Min, Y. L., Bassel-Duby, R. & Olson, E. N. CRISPR correction of duchenne muscular dystrophy. Annu. Rev. Med. 70, 239–255 (2019).
Ceccaldi, R., Rondinelli, B. & D’Andrea, A. D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64 (2016).
Zafra, M. P. et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 36, 888–893 (2018).
Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019).
Issa, J. P. Epigenetic variation and cellular Darwinism. Nat. Genet. 43, 724–726 (2011).
Zhu, M. et al. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell 177, 608–621 (2019). This study reports that somatic mutations accumulate in the liver and lead to increased hepatic clonal fitness in a context of chronic liver disease. This work demonstrates that SGR also exists in non-Mendelian disease.
Jacobsen, S. E. W. & Nerlov, C. Haematopoiesis in the era of advanced single-cell technologies. Nat. Cell Biol. 21, 2–8 (2019). This review describes the unanticipated plasticity and heterogeneity of HSCs.
Arredondo-Vega, F. X. et al. Adenosine deaminase deficiency with mosaicism for a ‘second-site suppressor’ of a splicing mutation: decline in revertant T lymphocytes during enzyme replacement therapy. Blood 99, 1005–1013 (2002).
Rieux-Laucat, F. et al. Inherited and somatic CD3ζ mutations in a patient with T cell deficiency. N. Engl. J. Med. 354, 1913–1921 (2006).
Fuchs, S. et al. Omenn syndrome associated with a functional reversion due to a somatic second-site mutation in CARD11 deficiency. Blood 126, 1658–1669 (2015).
Xia, B. et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 39, 159–161 (2007).
Wada, T. et al. Oligoclonal expansion of T lymphocytes with multiple second-site mutations leads to Omenn syndrome in a patient with RAG1-deficient severe combined immunodeficiency. Blood 106, 2099–2101 (2005). This study demonstrates that SGR can lead to deleterious consequences by generating pathogenic T lymphocytes and by causing Omenn syndrome.
Crescenzi, B. et al. Totipotent stem cells bearing del(20q) maintain multipotential differentiation in Shwachman Diamond syndrome. Br. J. Haematol. 144, 116–119 (2009).
Ariga, T., Yamada, M., Sakiyama, Y. & Tatsuzawa, O. A case of Wiskott–Aldrich syndrome with dual mutations in exon 10 of the WASP gene: an additional de novo one-base insertion, which restores frame shift due to an inherent one-base deletion, detected in the major population of the patient’s peripheral blood lymphocytes. Blood 92, 699–701 (1998).
Acknowledgements
Despite efforts to be as exhaustive as possible, the authors apologize to all colleagues whose work is not cited because of space limitations. P.R. warmly thanks J.P. de Villartay and S. Latour for discussions, advice and support, and also thanks the members of the DGSI laboratory. Current work in P.R.’s laboratory is funded by INSERM, Ligue contre le Cancer and INCa. P.R. is a staff scientist at the Centre National de la Recherche Scientifique (CNRS).
Peer review information
Nature Reviews Genetics thanks F. Candotti, D. Schindler, H.S. Scott and P. Venugopal for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
P.R. and A.F. contributed to all aspects of the article. C.K. reviewed the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Somatic mosaicism
-
Occurs when an individual contains two or more genetically distinct cellular populations that initially arose from the same fertilized egg.
- Biallelic mutations
-
Two (possibly different) mutations located on both alleles of the same gene.
- Interstitial deletions
-
Losses of an internal part of a chromosome.
- Chromothripsis
-
Complex genomic modification characterized by a high number of chromosomal rearrangements and copy number changes in one or a small number of chromosomes.
- Compound heterozygosity
-
The existence of distinct mutations on opposite alleles of a single gene.
- AluY motifs
-
Short, interspersed and repeated sequences integrated into the genome.
- Genomic instability
-
Increased rate of genomic mutation.
- Uniparental disomy
-
Occurs when both copies of a chromosome originate from one parent (maternal or paternal) and the chromosome from the other parent is absent. Segmental uniparental disomy occurs when only part of a chromosome is affected.
- Aneuploidy
-
An abnormal chromosome number resulting from gain or a loss of one or a few chromosomes or arms of a chromosome.
Rights and permissions
About this article
Cite this article
Revy, P., Kannengiesser, C. & Fischer, A. Somatic genetic rescue in Mendelian haematopoietic diseases. Nat Rev Genet 20, 582–598 (2019). https://doi.org/10.1038/s41576-019-0139-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-019-0139-x
This article is cited by
-
Human embryonic genetic mosaicism and its effects on development and disease
Nature Reviews Genetics (2024)
-
Genetic variation across and within individuals
Nature Reviews Genetics (2024)
-
Adaptive and Maladaptive Clonal Hematopoiesis in Telomere Biology Disorders
Current Hematologic Malignancy Reports (2024)
-
Somatic genetic variation in healthy tissue and non-cancer diseases
European Journal of Human Genetics (2023)
-
Genetics of human telomere biology disorders
Nature Reviews Genetics (2023)