Abstract
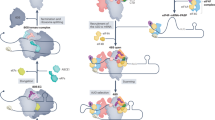
Regulation of mRNA translation offers the opportunity to diversify the expression and abundance of proteins made from individual gene products in cells, tissues and organisms. Emerging evidence has highlighted variation in the composition and activity of several large, highly conserved translation complexes as a means to differentially control gene expression. Heterogeneity and specialized functions of individual components of the ribosome and of the translation initiation factor complexes eIF3 and eIF4F, which are required for recruitment of the ribosome to the mRNA 5′ untranslated region, have been identified. In this Review, we summarize the evidence for selective mRNA translation by components of these macromolecular complexes as a means to dynamically control the translation of the proteome in time and space. We further discuss the implications of this form of gene expression regulation for a growing number of human genetic disorders associated with mutations in the translation machinery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kong, J. & Lasko, P. Translational control in cellular and developmental processes. Nat. Rev. Genet. 13, 383–394 (2012).
Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009).
Curtis, D., Lehmann, R. & Zamore, P. Translational regulation in development. Cell 81, 171–178 (1995).
Schwanhausser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
Fortelny, N., Overall, C. M., Pavlidis, P. & Freue, G. V. C. Can we predict protein from mRNA levels? Nature 547, E19–E20 (2017).
Floor, S. N. & Doudna, J. A. Tunable protein synthesis by transcript isoforms in human cells. eLife 5, e10921 (2016).
Ingolia, N. T. Ribosome footprint profiling of translation throughout the genome. Cell 165, 22–33 (2016).
Liu, T. Y. et al. Time-resolved proteomics extends ribosome profiling-based measurements of protein synthesis dynamics. Cell Syst. 4, 636–644 (2017).
Jovanovic, M. et al. Dynamic profiling of the protein life cycle in response to pathogens. Science 347, 1259038 (2015).
Khan, Z. et al. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 342, 1100–1104 (2013).
Fujii, K., Shi, Z., Zhulyn, O., Denans, N. & Barna, M. Pervasive translational regulation of the cell signalling circuitry underlies mammalian development. Nat. Commun. 8, 14443 (2017).
Kondrashov, N. et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397 (2011).
Slaidina, M. & Lehmann, R. Translational control in germline stem cell development. J. Cell Biol. 207, 13–21 (2014).
Sampath, P. et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2, 448–460 (2008).
Zhang, Q., Shalaby, N. A. & Buszczak, M. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science 343, 298–301 (2014).
Signer, R. A. J., Magee, J. A., Salic, A. & Morrison, S. J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014).
Shi, Z. et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 67, 71–83 (2017). This study uses absolute quantification mass spectrometry techniques to calculate the stoichiometry of several RPs on the ribosome, revealing heterogeneity in ribosome composition within mES cells.
Crick, F. H. C. On protein synthesis. Symp. Soc. Exp. Biol. 12, 138–163 (1958).
Crick, F. H. C. & Brenner, S. Some footnotes on protein synthesis: a note for the RNA Tie Club. Profiles in Medicine https://profiles.nlm.nih.gov/ps/access/SCBBFV.pdf (1959).
Brenner, S. & Crick, F. H. C. What are the properties of genetic RNA? A note for the RNA Tie Club. Profiles in Medicine https://profiles.nlm.nih.gov/ps/access/SCBBFZ.pdf (1960).
Jacob, F. & Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 (1961).
Brenner, S., Jacob, F. & Meselson, M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190, 576–581 (1961).
Lamfrom, H. Factors determining the specificity of hemoglobin synthesized in a cell-free system. J. Mol. Biol. 3, 241–252 (1961).
Marygold, S. J., Coelho, C. M. A. & Leevers, S. J. Genetic analysis of RpL38 and RpL5, two minute genes located in the centric heterochromatin of chromosome 2 of Drosophila melanogaster. Genetics 169, 683–695 (2005).
Guimaraes, J. C. & Zavolan, M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 17, 236 (2016). This study shows distinct RP expression signatures at the RNA level in particular cell types, most notably in the haematopoietic lineage. The observed patterns in RP expression may be regulated by cell type-specific transcription factors.
Ramagopal, S. Induction of cell-specific ribosomal proteins in aggregation-competent nonmorphogenetic Dictyostelium discoideum. Biochem. Cell Biol. 68, 1281–1287 (1990).
Lopes, A. M. et al. The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol. Biol. 11, 33 (2010).
Zhang, Y. et al. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev. Cell 24, 411–425 (2013). This study reveals distinct and antagonistic developmental functions for two RP paralogues, RPL22/eL22 and RPL22L1/eL22L1.
Xue, S. & Barna, M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369 (2012).
Byrgazov, K., Vesper, O. & Moll, I. Ribosome heterogeneity: another level of complexity in bacterial translation regulation. Curr. Opin. Microbiol. 16, 133–139 (2013).
Melnikov, S. et al. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567 (2012).
Noller, H. F., Hoffarth, V. & Zimniak, L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256, 1416–1419 (1992).
Anger, A. M. et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013).
Leppek, K., Das, R. & Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 19, 158–174 (2018).
Simsek, D. et al. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169, 1051–1065 (2017). This study uses quantitative mass spectrometry techniques to identify hundreds of RAPs.
Anderson, L. & Hunter, C. L. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteom. 5, 573–588 (2006).
Picotti, P. & Aebersold, R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566 (2012).
Addona, T. A. et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 (2009).
Kusebauch, U. et al. Human SRMAtlas: a resource of targeted assays to quantify the complete human proteome. Cell 166, 766–778 (2016).
Peterson, A. C., Russell, J. D., Bailey, D. J., Westphall, M. S. & Coon, J. J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteom. 11, 1475–1488 (2012).
van de Waterbeemd, M. et al. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods 14, 283–286 (2017). This study improves native mass spectrometry techniques to provide sufficient resolution to identify heterogeneity in prokaryotic ribosomes.
Slavov, N., Semrau, S., Airoldi, E., Budnik, B. & van Oudenaarden, A. Differential stoichiometry among core ribosomal proteins. Cell Rep. 13, 865–873 (2015).
Gupta, V. & Warner, J. R. Ribosome-omics of the human ribosome. RNA 20, 1004–1013 (2014).
Ishii, K. et al. Characteristics and clustering of human ribosomal protein genes. BMC Genomics 7, 37 (2006).
Bortoluzzi, S., D’Alessi, F., Romualdi, C. & Danieli, G. A. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics 17, 1152–1157 (2001).
Wong, Q. W.-L. et al. RPL39L is an example of a recently evolved ribosomal protein paralog that shows highly specific tissue expression patterns and is upregulated in ESCs and HCC tumors. RNA Biol. 11, 33–41 (2014).
De Klerk, E. et al. Assessing the translational landscape of myogenic differentiation by ribosome profiling. Nucleic Acids Res. 43, 4408–4428 (2015).
Sugihara, Y. et al. Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J. Proteome Res. 9, 1351–1366 (2010).
Chaillou, T., Zhang, X. & Mccarthy, J. J. Expression of muscle-specific ribosomal protein L3-like impairs myotube growth. J. Cell. Physiol. 231, 1894–1902 (2016).
Perry, R. P. The architecture of mammalian ribosomal protein promoters. BMC Evol. Biol. 5, 15 (2005).
Macias, S., Bragulat, M., Tardiff, D. F. & Vilardell, J. L30 binds the nascent RPL30 transcript to repress U2 snRNP recruitment. Mol. Cell 30, 732–742 (2008).
Malygin, A. A., Parakhnevitch, N. M., Ivanov, A. V., Eperon, I. C. & Karpova, G. G. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 35, 6414–6423 (2007).
Ivanov, A. V., Malygin, A. a. & Karpova, G. G. Human ribosomal protein S26 suppresses the splicing of its pre-mRNA. Biochim. Biophys. Acta 1727, 134–140 (2005).
O’Leary, M. N. et al. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 9, e1003708 (2013).
Reuveni, S., Ehrenberg, M. & Paulsson, J. Ribosomes are optimized for autocatalytic production. Nature 547, 293–297 (2017).
Lam, Y. W., Lamond, A. I., Mann, M. & Andersen, J. S. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 (2007).
Mazumder, B. et al. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115, 187–198 (2003).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Sharma, S. & Lafontaine, D. L. J. ‘View from a bridge’: a new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 40, 560–575 (2015).
Mcmahon, M., Contreras, A. & Ruggero, D. Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip. Rev. RNA 6, 173–189 (2015).
Bellodi, C. et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Rep. 3, 1493–1502 (2013).
Yoon, A. et al. Impaired control of IRES-mediated translation in X-linked Dyskeratosis Congenita. Science 312, 902–907 (2006).
Locati, M. D. et al. Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA 23, 1188–1199 (2017). This paper reveals a switch in ribosome composition, from an embryonic-specific rRNA allele to a somatic rRNA with a different sequence, during zebrafish development.
Locati, M. D. et al. Linking maternal and somatic 5S rRNA types with different sequence-specific non-LTR retrotransposons. RNA 23, 446–456 (2017).
Arnheim, N. & Southern, E. M. Heterogeneity of the ribosomal genes in mice and men. Cell 11, 363–370 (1977).
Parks, M. M. et al. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 4, eaao0665 (2018).
Kuo, B. A., Gonzalez, I. L., Gillespie, D. A. & Sylvester, J. E. Human ribosomal RNA variants from a single individual and their expression in different tissues. Nucleic Acids Res. 24, 4817–4824 (1996).
Landry, D. M., Hertz, M. I. & Thompson, S. R. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 23, 2753–2764 (2009).
Hertz, M. I., Landry, D. M., Willis, A. E., Luo, G. & Thompson, S. R. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell. Biol. 33, 1016–1026 (2013).
Quade, N., Boehringer, D., Leibundgut, M., van den Heuvel, J. & Ban, N. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat. Commun. 6, 7646 (2015).
Nishiyama, T., Yamamoto, H., Uchiumi, T. & Nakashima, N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 35, 1514–1521 (2007).
Boria, I. et al. The ribosomal basis of diamond-blackfan anemia: mutation and database update. Hum. Mutat. 31, 1269–1279 (2010).
Bolze, A. et al. Haploinsufficiency in humans with isolated congenital asplenia. Science 340, 976–978 (2013).
Klauck, S. M. et al. Mutations in the ribosomal protein gene RPL10 suggest a novel modulating disease mechanism for autism. Mol. Psychiatry 11, 1073–1084 (2006).
Zhou, C. et al. Mutation in ribosomal protein L21 underlies hereditary hypotrichosis simplex. Hum. Mutat. 32, 710–714 (2011).
Oliver, E. R., Saunders, T. L., Tarle, S. A. & Glaser, T. Ribosomal protein L24 defect in Belly spot and tail (Bst), a mouse Minute. Development 131, 3907–3920 (2004).
Anderson, S. J. et al. Ablation of ribosomal protein L22 selectively impairs αβ T cell development by activation of a p53-dependent checkpoint. Immunity 26, 759–772 (2007).
Zhang, Y. et al. Ribosomal proteins Rpl22 and Rpl22l1 control morphogenesis by regulating pre-mRNA splicing. Cell Rep. 18, 545–556 (2017).
Mills, E. W. & Green, R. Ribosomopathies: there’s strength in numbers. Science 358, eaan2755 (2017).
Brotherton, T. W., Chui, D. H. K., McFarland, E. C. & Russell, E. S. Fetal erythropoiesis and hemoglobin ontogeny in tail-short (Ts/+) mutant mice. Blood 54, 673–683 (1979).
Zhang, Y. & Lu, H. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377 (2009).
Xue, S. et al. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38 (2015).
Horos, R. et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood 119, 262–272 (2012).
Majzoub, K. et al. RACK1 controls IRES-mediated translation of viruses. Cell 159, 1086–1095 (2014).
Kadrmas, J. L., Smith, M. A., Pronovost, S. M. & Beckerle, M. C. Characterization of RACK1 function in Drosophila development. Dev. Dyn. 236, 2207–2215 (2007).
Volta, V. et al. RACK1 depletion in a mouse model causes lethality, pigmentation deficits and reduction in protein synthesis efficiency. Cell. Mol. Life Sci. 70, 1439–1450 (2013).
Lee, A. S., Burdeinick-Kerr, R. & Whelan, S. P. J. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc. Natl Acad. Sci. USA 110, 324–329 (2013).
Dobbelstein, M. & Shenk, T. In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J. Virol. 69, 8027–8034 (1995).
Ludwig, L. S. et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat. Med. 20, 748–753 (2014).
Komatsu, M. et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 23, 1977–1986 (2004).
Zhang, M. et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 22, 1922–1934 (2015).
Dong, J. et al. Rps3/uS3 promotes mRNA binding at the 40S ribosome entry channel and stabilizes preinitiation complexes at start codons. Proc. Natl Acad. Sci. USA 114, e2126–e2135 (2017).
Darnell, J. C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Chen, E., Sharma, M. R., Shi, X., Agrawal, R. K. & Joseph, S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell 54, 407–417 (2014).
Vasilyev, N. et al. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc. Natl Acad. Sci. USA 112, E5391–E5400 (2015).
Meyer, K. D. et al. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015). This study reveals that m 6 A methylation in the 5′ UTR promotes cap-independent translation initiation via the eIF3 complex.
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Jackson, R. J., Hellen, C. U. T. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 10, 113–127 (2010).
Hinnebusch, A. G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812 (2014).
Mohammad, M. P., Munzarová Pondelícková, V., Zeman, J., Gunišová, S. & Valášek, L. S. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res. 45, 2658–2674 (2017).
Cuchalová, L. et al. The RNA recognition motif of eukaryotic translation initiation factor 3 g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol. 30, 4671–4686 (2010).
Pöyry, T. A. A., Kaminski, A. & Jackson, R. J. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 18, 62–75 (2004).
Hinnebusch, A. G. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31, 553–562 (2006).
Masutani, M., Sonenberg, N., Yokoyama, S. & Imataka, H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 26, 3373–3383 (2007).
Wagner, S., Herrmannová, A., Malík, R., Peclinovská, L. & Valášek, L. S. Functional and biochemical characterization of human eukaryotic translation initiation factor 3 in living cells. Mol. Cell. Biol. 34, 3041–3052 (2014).
ElAntak, L. et al. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J. Mol. Biol. 396, 1097–1116 (2010).
Damoc, E. et al. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell. Proteom. 6, 1135–1146 (2007). This study performs both bottom-up and top-down proteomics methods to identify PTMs on the eIF3 complex.
Jia, W., Andaya, A. & Leary, J. A. Novel mass spectrometric method for phosphorylation quantification using cerium oxide nanoparticles and tandem mass tags. Anal. Chem. 84, 2466–2473 (2010).
Andaya, A., Villa, N., Jia, W., Fraser, C. S. & Leary, J. A. Phosphorylation stoichiometries of human Eukaryotic initiation factors. Int. J. Mol. Sci. 15, 11523–11538 (2014).
Cai, Q. et al. Distinct regions of human eIF3 are sufficient for binding to the HCV IRES and the 40S ribosomal subunit. J. Mol. Biol. 403, 185–196 (2010).
Hashem, Y. et al. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503, 539–543 (2013).
Siridechadilok, B., Fraser, C. S., Hall, R. J., Doudna, J. A. & Nogales, E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310, 1513–1515 (2005).
Lee, A. S. Y., Kranzusch, P. J. & Cate, J. H. D. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114 (2015).
Lee, A. S., Kranzusch, P. J., Doudna, J. A. & Cate, J. H. D. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99 (2016).
Sonenberg, N., Morgan, M. A., Testa, D., Colonno, R. J. & Shatkin, A. J. Interaction of a limited set of proteins with different mRNAs and protection of 5′ caps against pyrophosphatase digestion in initiation complexes. Nucleic Acids Res. 7, 15–29 (1979).
Kumar, P., Hellen, C. U. T. & Pestova, T. V. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 30, 1573–1588 (2016).
Zhang, L., Pan, X. & Hershey, J. W. B. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J. Biol. Chem. 282, 5790–5800 (2007).
Hershey, J. W. B. The role of eIF3 and its individual subunits in cancer. Biochim. Biophys. Acta - Gene Regul. Mech. 1849, 792–800 (2015).
Choudhuri, A., Evans, T. & Maitra, U. Non-core subunit eIF3h of translation initiation factor eIF3 regulates zebrafish embryonic development. Dev. Dyn. 239, 1632–1644 (2010).
Choudhuri, A., Maitra, U. & Evans, T. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proc. Natl Acad. Sci. USA 110, 9818–9823 (2013). This paper identifies a developmental function for a subunit of eIF3 in the zebrafish eye. Knockdown of the subunit decreased translation of multiple mRNAs, including those encoding proteins required for proper eye formation.
Zhou, F., Roy, B. & von Arnim, A. G. Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol. 10, 193 (2010).
Kim, T., Kim, B., Yahalom, A., Chamovitz, D. A. & von Arnim, A. G. Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16, 3341–3356 (2004).
Daxinger, L. et al. A forward genetic screen identifies eukaryotic translation initiation factor 3, subunit H (eIF3h), as an enhancer of variegation in the mouse. G3 2, 1393–1396 (2012).
Dhalia, R. et al. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol. Biochem. Parasitol. 140, 23–41 (2005).
Freire, E. R. et al. The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol. Biochem. Parasitol. 176, 25–36 (2011).
Moura, D. M. N. et al. Two related trypanosomatid eIF4G homologues have functional differences compatible with distinct roles during translation initiation. RNA Biol. 12, 305–319 (2015).
Joshi, B., Cameron, A. & Jagus, R. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 271, 2189–2203 (2004).
Landon, A. L. et al. MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nat. Commun. 5, 5413 (2014).
Sugiyama, H. et al. Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 114, 340–345 (2017).
Caron, S., Charon, M., Cramer, E., Sonenberg, N. & Dusanter-Fourt, I. Selective modification of eukaryotic initiation factor 4F (eIF4F) at the onset of cell differentiation: recruitment of eIF4GII and long-lasting phosphorylation of eIF4E. Mol. Cell. Biol. 24, 4920–4928 (2004).
Hernández, G. & Vazquez-Pianzola, P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech. Dev. 122, 865–876 (2005).
Tee, A. R., Tee, J. A. & Blenis, J. Characterizing the interaction of the mammalian eIF4E-related protein 4EHP with 4E-BP1. FEBS Lett. 564, 58–62 (2004).
Ho, J. J. D. & Lee, S. A. Cap for every occasion: alternative eIF4F complexes. Trends Biochem. Sci. 41, 821–823 (2016).
Waskiewicz, A. J., Flynn, A., Proud, C. G. & Cooper, J. A. Mitogen-activated protein kinases activate the serine/threonine kinaseses Mnk1 and Mnk2. EMBO J. 16, 1909–1920 (1997).
Dobrikov, M., Dobrikova, E., Shveygert, M. & Gromeier, M. Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase Cα regulates eIF4G1 binding to Mnk1. Mol. Cell. Biol. 31, 2947–2959 (2011).
Raught, B. et al. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 19, 434–444 (2000).
Dobrikov, M. I., Shveygert, M., Brown, M. C. & Gromeier, M. Mitotic phosphorylation of eukaryotic initiation factor 4G1 (eIF4G1) at Ser1232 by Cdk1:Cyclin B inhibits eIF4A helicase complex binding with RNA. Mol. Cell. Biol. 34, 439–451 (2014).
Xu, X., Vatsyayan, J., Gao, C., Bakkenist, C. J. & Hu, J. Sumoylation of eIF4E activates mRNA translation. EMBO Rep. 11, 299–304 (2010).
Jongjitwimol, J. et al. The S. pombe translation initiation factor eIF4G is sumoylated and associates with the SUMO protease Ulp2. PLoS ONE 9, e94182 (2014).
Sun, F., Palmer, K. & Handel, M. A. Mutation of Eif4g3, encoding a eukaryotic translation initiation factor, causes male infertility and meiotic arrest of mouse spermatocytes. Development 137, 1699–1707 (2010).
Ghosh, S. & Lasko, P. Loss-of-function analysis reveals distinct requirements of the translation initiation factors eIF4E, eIF4E-3, eIF4G and eIF4G2 in Drosophila spermatogenesis. PLoS ONE 10, e0122519 (2015).
Contreras, V., Richardson, M.a, Hao, E. & Keiper, B. D. Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death Differ. 15, 1232–1242 (2008).
Friday, A. J. & Keiper, B. D. Positive mRNA translational control in germ cells by initiation factor selectivity. Biomed. Res. Int. 2015, 327963 (2015).
Morita, M. et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell. Biol. 32, 3585–3593 (2012).
Duncan, R., Milburn, S. C. & Hershey, J. W. B. Regulated phosphorylation and low abundance of HeLa cell initiation factor 4F suggest a role in translational control. Heat shock effect on eIF4F. J. Biol. Chem. 262, 380–388 (1987).
Truitt, M. L. et al. Differential requirements for eIF4E dose in normal development and cancer. Cell 162, 59–71 (2015). This study presents an eIF4E haploinsufficiency mouse model that reveals that eIF4E expression is in excess for normal development but not for cancer formation and that eIF4E is required for translation of specific mRNAs via a novel 5′ UTR element.
Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012).
Levy, S., Avni, D., Hariharan, N., Perry, R. P. & Meyuhas, O. Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc. Natl Acad. Sci. USA 88, 3319–3323 (1991).
Osborne, M. J. et al. eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition. Proc. Natl Acad. Sci. USA 110, 3877–3882 (2013).
Ueda, T., Watanabe-Fukunaga, R., Fukuyama, H., Nagata, S. & Fukunaga, R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 24, 6539–6549 (2004).
Furic, L. et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl Acad. Sci. USA 107, 14134–14139 (2010).
Moy, J. K. et al. The MNK-eIF4E signaling axis contributes to injury-induced nociceptive plasticity and the development of chronic pain. J. Neurosci. 37, 7841–7499 (2017).
Connolly, E., Braunstein, S., Formenti, S. & Schneider, R. J. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol. Cell. Biol. 26, 3955–3965 (2006).
Timpano, S. & Uniacke, J. Human cells cultured under physiological oxygen utilize two cap-binding proteins to recruit distinct mRNAs for translation. J. Biol. Chem. 291, 10772–10782 (2016).
Ho, J. J. D. et al. Systemic reprogramming of translation efficiencies on oxygen stimulus. Cell Rep. 14, 1293–1300 (2016).
Braunstein, S. et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell 28, 501–512 (2007).
Clemens, M. J., Bushell, M. & Morley, S. J. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene 17, 2921–2931 (1998).
Marissen, W. E. & Lloyd, R. E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 18, 7565–7574 (1998).
Bushell, M. et al. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death Differ. 7, 628–636 (2000).
Nevins, T. A., Harder, Z. M., Korneluk, R. G. & Holcík, M. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 278, 3572–3579 (2003).
Henis-Korenblit, S. et al. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl Acad. Sci. USA 99, 5400–5405 (2002).
Hundsdoerfer, P., Thoma, C. & Hentze, M. W. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl Acad. Sci. USA 102, 13421–13426 (2005).
Ramirez-Valle, F., Braunstein, S., Zavadil, J., Formenti, S. C. & Schneider, R. J. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 181, 293–307 (2008).
Garzia, A. et al. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun. 8, 16056 (2017).
De Benedetti, A. & Graff, J. R. eIF-4E expression and its role in malignancies and metastases. Oncogene 23, 3189–3199 (2004).
Pelletier, J., Graff, J., Ruggero, D. & Sonenberg, N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 75, 250–263 (2015).
Howard, A. & Rogers, A. N. Role of translation initiation factor 4G in lifespan regulation and age-related health. Ageing Res. Rev. 13, 115–124 (2014).
Blagden, S. P. & Willis, A. E. The biological and therapeutic relevance of mRNA translation in cancer. Nat. Rev. Clin. Oncol. 8, 280–291 (2011).
Poss, Z. C., Ebmeier, C. C. & Taatjes, D. J. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 48, 575–608 (2013).
Villani, A. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Trapnell, C. Defining cell types and states with single-cell genomics. Genome Res. 25, 1491–1498 (2015).
Hughes, A. J. et al. Single-cell western blotting. Nat. Methods 11, 749–755 (2014).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Moor, A. E. et al. Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science 2399, 1299–1303 (2017).
Morisaki, T. et al. Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429 (2016).
Wu, B., Eliscovich, C., Yoon, Y. J. & Singer, R. H. Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435 (2016).
Yan, X., Hoek, T. A., Vale, R. D. & Tanenbaum, M. E. Dynamics of translation of single mRNA molecules in vivo. Cell 165, 976–989 (2016).
Han, B. G., Watson, Z., Cate, J. H. D. & Glaeser, R. M. Monolayer-crystal streptavidin support films provide an internal standard of cryo-EM image quality. J. Struct. Biol. 200, 307–313 (2017).
des Georges, A. et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525, 491–495 (2015).
Fraser, C. S., Berry, K. E., Hershey, J. W. B. & Doudna, J. A. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol. Cell 26, 811–819 (2007).
Villa, N., Do, A., Hershey, J. W. B. & Fraser, C. S. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem. 288, 32932–32940 (2013).
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011).
Fraser, C. S. & Doudna, J. A. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 5, 29–38 (2007).
Weingarten-Gabbay, S. et al. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351, aad4939 (2016).
Archer, S. K., Shirokikh, N. E., Beilharz, T. H. & Preiss, T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535, 570–574 (2016).
Simsek, D. & Barna, M. An emerging role for the ribosome as a nexus for post-translational modifications. Curr. Opin. Cell Biol. 45, 92–101 (2017).
Sharma, D., Cukras, A. R., Rogers, E. J., Southworth, D. R. & Green, R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J. Mol. Biol. 374, 1065–1076 (2007).
Loenarz, C. et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl Acad. Sci. USA 111, 4019–4024 (2014).
Paolini, N. A. et al. A ribosomopathy reveals decoding defective ribosomes driving human dysmorphism. Am. J. Hum. Genet. 100, 506–522 (2017).
Singleton, R. S. et al. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc. Natl Acad. Sci. USA 111, 4031–4036 (2014).
Jha, S. et al. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 546, 651–655 (2017).
Finley, D., Bartel, B. & Varshavsky, A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338, 394–401 (1989).
Kobayashi, M. et al. The ubiquitin hybrid gene UBA52 regulates ubiquitination of ribosome and sustains embryonic development. Sci. Rep. 6, 36780 (2016).
Higgins, R. et al. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol. Cell 59, 35–49 (2015).
Sung, M. K. et al. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. eLife 5, e19105 (2016).
Nguyen, A. T. et al. UBE2O remodels the proteome during terminal erythroid differentiation. Science 357, eaan0218 (2017).This paper reveals extensive ubiquitylation of several RPs by a ubiquitin ligase, UBE2O, which is required for erythropoiesis.
Yanagitani, K., Juszkiewicz, S. & Hegde, R. S. UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357, 472–475 (2017).
Xirodimas, D. P. et al. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 9, 280–286 (2008).
Matafora, V., D’Amato, A., Mori, S., Blasi, F. & Bachi, A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol. Cell. Proteom. 8, 2243–2255 (2009).
Panse, V. G. et al. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 7, 1311–1321 (2006).
Sun, C. et al. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc. Natl Acad. Sci. USA 108, 20473–20478 (2011).
Panic, L. et al. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol. Cell. Biol. 26, 8880–8891 (2006).
McGowan, K. A. et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 40, 963–970 (2008).
Mcgowan, K. A. et al. Reduced ribosomal protein gene dosage and p53 activation in low-risk myelodysplastic syndrome. Blood 118, 3622–3633 (2011).
Sulic, S. et al. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 19, 3070–3082 (2005).
Gazda, H. T. et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan Anemia patients. Am. J. Hum. Genet. 83, 769–780 (2008).
Watkins-Chow, D. E. et al. Mutation of the Diamond-Blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet. 9, e1003094 (2013).
Doherty, L. et al. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 86, 222–228 (2010).
Ebert, B. L. et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451, 335–339 (2008).
Barlow, J. L. et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat. Med. 16, 59–66 (2010).
Ikeda, F. et al. Exome sequencing identified RPS15A as a novel causative gene for Diamond-Blackfan anemia. Haematologica 102, e93–e96 (2017).
Cmejla, R., Cmejlova, J., Handrkova, H., Petrak, J. & Pospisilova, D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum. Mutat. 28, 1178–1182 (2007).
Song, M. et al. A novel initiation codon mutation in the ribosomal protein S17 gene (RPS17) in a patient with Diamond-Blackfan anemia. Pediatr. Blood Cancer 54, 629–631 (2010).
Draptchinskaia, N. et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 21, 169–175 (1999).
Gazda, H. T. et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 79, 1110–1118 (2006).
Farrar, J. E. et al. Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood 118, 6943–6951 (2011).
Ferretti, M. B., Ghalei, H., Ward, E. A., Potts, E. L. & Karbstein, K. Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements. Nat. Struct. Mol. Biol. 24, 700–707 (2017).
Wang, R. et al. Loss of function mutations in RPL27 and RPS27 identified by whole-exome sequencing in Diamond-Blackfan anaemia. Br. J. Haematol. 168, 854–864 (2015).
Gripp, K. W. et al. Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am. J. Med. Genet. Part A 164, 2240–2249 (2014).
Mirabello, L. et al. Whole-exome sequencing and functional studies identify RPS29 as a novel gene mutated in multicase Diamond-Blackfan anemia families. Blood 124, 24–32 (2014).
Gu, Y. A. N. et al. Deficiency of monoclonal non-specific suppressor factor beta (MNSFB) promotes pregnancy loss in mice. Mol. Reprod. Dev. 82, 475–488 (2015).
Perucho, L. et al. RPLP1, a crucial ribosomal protein for embryonic development of the nervous system. PLoS ONE 9, e99956 (2014).
Brooks, S. S. et al. A novel ribosomopathy caused by dysfunction of RPL10 disrupts neurodevelopment and causes X-linked microcephaly in humans. Genetics 198, 723–733 (2014).
Morgado-Palacin, L. et al. Partial loss of Rpl11 in adult mice recapitulates Diamond-Blackfan anemia and promotes lymphomagenesis. Cell Rep. 13, 712–722 (2015).
Poddar, D. et al. An extraribosomal function of ribosomal protein L13a in macrophages resolves inflammation. J. Immunol. 190, 3600–3612 (2013).
Chaudhuri, S. et al. Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA 13, 2224–2237 (2007).
Landowski, M. et al. Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Hum. Genet. 132, 1265–1274 (2013).
Rao, S. et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 120, 3764–3773 (2012).
Gazda, H. T. et al. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in Diamond-Blackfan anemia. Hum. Mutat. 33, 1037–1044 (2012).
Terzian, T. et al. Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models. J. Pathol. 224, 540–552 (2011).
Oristian, D. S. et al. Ribosomal protein L29 / HIP deficiency delays osteogenesis and increases fragility of adult bone in mice. J. Orthop. Res. 27, 28–35 (2009).
Kirn-Safran, C. B. et al. Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev. Dyn. 236, 447–460 (2007).
Farrar, J. E. et al. Exploiting pre-rRNA processing in Diamond Blackfan anemia gene discovery and diagnosis. Am. J. Hematol. 89, 985–991 (2014).
Farrar, J. E. et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood 112, 1582–1592 (2008).
Acknowledgements
The authors thank members of the Barna laboratory for thoughtful comments on this work. This work was supported by the New York Stem Cell Foundation NYSCF-R-I36 (M.B.), National Institutes of Health grant 1R01HD086634 (M.B.) and Pew Scholars Award (M.B.). N.R.G. is supported by National Science Foundation Graduate Research Fellowship DGE-114747. M.B. is a New York Stem Cell Foundation Robertson Investigator.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Genetics thanks J. McCarthy, T. Preiss and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Glossary
- Ribosome
-
The macromolecular machine that synthesizes proteins, consisting of a large (60S) and a small (40S) subunit that come together to form the translationally active eukaryotic 80S ribosome.
- Start codon
-
The codon where translation is initiated, encoding the first amino acid of the peptide; typically an AUG encoding methionine.
- Kozak sequence
-
The optimal sequence context for a start codon.
- Open reading frame
-
(ORF). A region of mRNA, beginning with a start codon and ending with a stop codon, that has the capability of being translated into a peptide.
- Core ribosomal protein
-
(RP). One of the 80 canonical RPs, which were originally identified by their high-affinity association with the ribosome.
- Ribosomal RNA
-
(rRNA). The RNA components of the ribosome. Three eukaryotic rRNAs (5S, 5.8S and 28S) are in the large ribosomal subunit, while one (18S) is in the small subunit.
- Expansion segments
-
Sections of ribosomal RNA (rRNA) that are longer in eukaryotes relative to prokaryotes. Many of these expansion segments also show growth between unicellular and multicellular organisms.
- Untranslated region
-
(UTR). The portions of the mature mRNA that do not encode a protein. The sequence before the start codon is the 5′ UTR, while the section after the stop codon is the 3′ UTR.
- Paralogues
-
Genes, typically created by a duplication event, with high homology to another gene within the same species.
- Stoichiometry
-
The quantity of each constituent of a complex. If a subunit is not present on every complex, it is considered substoichiometric.
- Polysomes
-
Multiple ribosomes that are translating the same mRNA transcript.
- Proximity labelling
-
A technique to identify proteins in a subcellular region of interest. A promiscuous biotinylating enzyme is localized to a particular organelle or fused to a protein of interest and labels any proteins in its vicinity with biotin. Streptavidin pull-down and mass spectrometry can be used to identify these protein interactors.
- Pseudogenes
-
A paralogue that can no longer be functionally expressed.
- Internal ribosome entry sites
-
(IRESs). Regions in mRNA with the capability of initiating translation by recruiting translation machinery independently of a 5′ cap.
- Ribosomopathies
-
Human diseases caused by ribosomal defects.
- Diamond–Blackfan anaemia
-
(DBA). A common ribosomopathy caused by mutations in ribosomal proteins that results in anaemia and a variety of congenital birth defects.
- Haploinsufficiency
-
When the expression of a gene from a single functional allele is not sufficient for normal cellular function.
- 5′ cap
-
A methylated guanine that is present at every 5′ end of all eukaryotic transcripts.
- Preinitiation complex
-
(PIC). The ribosome and associated initiation factors that come together at specific stages of translation initiation. The eukaryotic 43S PIC contains the ribosome, eIF3, eIF2, eIF1, eIF1A and the initiator tRNA; the 48S PIC has the addition of eIF4F.
- Translation efficiency
-
A measure of the rate of translation for a particular transcript of interest; typically calculated by normalizing ribosome occupancy to mRNA abundance.
Rights and permissions
About this article
Cite this article
Genuth, N.R., Barna, M. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat Rev Genet 19, 431–452 (2018). https://doi.org/10.1038/s41576-018-0008-z
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-018-0008-z
This article is cited by
-
Shared and distinct mechanisms of UBA1 inactivation across different diseases
The EMBO Journal (2024)
-
Translation dysregulation in neurodegenerative diseases: a focus on ALS
Molecular Neurodegeneration (2023)
-
A single pseudouridine on rRNA regulates ribosome structure and function in the mammalian parasite Trypanosoma brucei
Nature Communications (2023)
-
Salamanders’ regenerative potential might be driven by a specific protein variant
Nature (2023)
-
The Akt/mTOR and MNK/eIF4E pathways rewire the prostate cancer translatome to secrete HGF, SPP1 and BGN and recruit suppressive myeloid cells
Nature Cancer (2023)