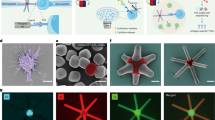
Abstract
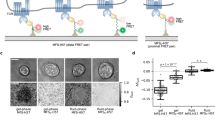
Elucidating the rules for receptor triggering in cell–cell and cell–matrix contacts requires precise control of ligand positioning in three dimensions. Here, we use the T cell receptor (TCR) as a model and subject T cells to different geometric arrangements of ligands, using a nanofabricated single-molecule array platform. This comprises monovalent TCR ligands anchored to lithographically patterned nanoparticle clusters surrounded by mobile adhesion molecules on a supported lipid bilayer. The TCR ligand could be co-planar with the supported lipid bilayer (2D), excluding the CD45 transmembrane tyrosine phosphatase, or elevated by 10 nm on solid nanopedestals (3D), allowing closer access of CD45 to engaged TCR. The two configurations resulted in different T cell responses, depending on the lateral spacing between the ligands. These results identify the important contributions of lateral and axial components of ligand positioning and create a more complete foundation for receptor engineering for immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Iwashima, M., Irving, B. A., van Oers, N. S., Chan, A. C. & Weiss, A. Sequential interactions of the TCR with two distinct cytoplasmic tyrosine kinases. Science 263, 1136–1139 (1994).
Liaunardy-Jopeace, A., Murton, B. L., Mahesh, M., Chin, J. W. & James, J. R. Encoding optical control in LCK kinase to quantitatively investigate its activity in live cells. Nat. Struct. Mol. Biol. 24, 1155–1163 (2017).
Sherman, E. et al. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity 35, 705–720 (2011).
Pageon, S. V. et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl Acad. Sci. USA 113, E5454–E5463 (2016).
Hui, E. & Vale, R. D. In vitro membrane reconstitution of the T-cell receptor proximal signaling network. Nat. Struct. Mol. Biol. 21, 133–142 (2014).
Chang, V. T. et al. Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol. 17, 574–582 (2016).
Davis, S. J. & van der Merwe, P. A. The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 7, 803–809 (2006).
Cochran, J. R., Cameron, T. O., Stone, J. D., Lubetsky, J. B. & Stern, L. J. Receptor proximity, not intermolecular orientation, is critical for triggering T-cell activation. J. Biol. Chem. 276, 28068–28074 (2001).
Deeg, J. et al. T cell activation is determined by the number of presented antigens. Nano Lett. 13, 5619–5626 (2013).
Matic, J., Deeg, J., Scheffold, A., Goldstein, I. & Spatz, J P. Fine tuning and efficient T cell activation with stimulatory aCD3 nanoarrays. Nano Lett. 13, 5090–5097 (2013).
Delcassian, D. et al. Nanoscale ligand spacing influences receptor triggering in T cells and NK cells. Nano Lett. 13, 5608–5614 (2013).
Choudhuri, K., Wiseman, D., Brown, M. H., Gould, K. & van der Merwe, P. A. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature 436, 578–582 (2005).
Carbone, C. B. et al. In vitro reconstitution of T cell receptor-mediated segregation of the CD45 phosphatase. Proc. Natl Acad. Sci. USA 114, E9338–E9345 (2017).
Chen, B. M. et al. The affinity of elongated membrane-tethered ligands determines potency of T cell receptor triggering. Front. Immunol. 8, 793 (2017).
Irles, C. et al. CD45 ectodomain controls interaction with GEMs and Lck activity for optimal TCR signaling. Nat. Immunol. 4, 189–197 (2003).
James, J. R. & Vale, R. D. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature 487, 64–69 (2012).
Schmid, E. M. et al. Size-dependent protein segregation at membrane interfaces. Nat. Phys. 12, 704–711 (2016).
Wu, Y., Vendome, J., Shapiro, L., Ben-Shaul, A. & Honig, B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature 475, 510–513 (2011).
Schvartzman, M. & Wind, S. J. Robust pattern transfer of nanoimprinted features for sub-5-nm fabrication. Nano Lett. 9, 3629–3634 (2009).
Cai, H. et al. Molecular occupancy of nanodot arrays. ACS Nano 10, 4173–4183 (2016).
Mossman, K. D., Campi, G., Groves, J. T. & Dustin, M. L. Altered TCR signaling from geometrically repatterned immunological synapses. Science 310, 1191–1193 (2005).
Roman, G., Martin, M. & Joachim, P. S. Block copolymer micelle nanolithography. Nanotechnology 14, 1153 (2003).
Schoen, I., Hu, W., Klotzsch, E. & Vogel, V. Probing cellular traction forces by micropillar arrays: contribution of substrate warping to pillar deflection. Nano Lett. 10, 1823–1830 (2010).
Bettinger, C. J., Langer, R. & Borenstein, J. T. Engineering substrate topography at the micro- and nanoscale to control cell function. Angew. Chem. Int. Ed. 48, 5406–5415 (2009).
Vardhana, S., Choudhuri, K., Varma, R. & Dustin, M. L. Essential role of ubiquitin and TSG101 protein in formation and function of the central supramolecular activation cluster. Immunity 32, 531–540 (2010).
Cai, E. et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science 356, eaal3118 (2017).
Shi, X. et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature 493, 111–115 (2013).
Gagnon, E., Schubert, D. A., Gordo, S., Chu, H. H. & Wucherpfennig, K. W. Local changes in lipid environment of TCR microclusters regulate membrane binding by the CD3ε cytoplasmic domain. J. Exp. Med. 209, 2423–2439 (2012).
Minguet, S., Swamy, M., Alarcón, B., Luescher, I. F. & Schamel, W. W. A. Full activation of the T Cell receptor requires both clustering and conformational changes at CD3. Immunity 26, 43–54 (2007).
Tolar, P., Sohn, H. W. & Pierce, S. K. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat. Immunol. 6, 1168–1176 (2005).
Shah, N. H. et al. An electrostatic selection mechanism controls sequential kinase signaling downstream of the T cell receptor. eLife 5, e20105 (2016).
Su, X., Ditlev, J. A., Rosen, M. K. & Vale, R. D. Reconstitution of TCR signaling using supported lipid bilayers. Methods Mol. Biol. 1584, 65–76 (2017).
Stone, M. B., Shelby, S. A., Nunez, M. F., Wisser, K. & Veatch, S. L. Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. eLife 6, e19891 (2017).
Grakoui, A. et al. The immunological synapse: a molecular machine controlling T cell activation. Science 285, 221–227 (1999).
Lohmuller, T. et al. Supported membranes embedded with fixed arrays of gold nanoparticles. Nano Lett. 11, 4912–4918 (2011).
Manz, B. N., Jackson, B. L., Petit, R. S., Dustin, M. L. & Groves, J. T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc. Natl Acad. Sci. USA 108, 9089–9094 (2011).
Taylor, M. J., Husain, K., Gartner, Z. J., Mayor, S. & Vale, R. D. A DNA-based T cell receptor reveals a role for receptor clustering in ligand discrimination. Cell 169, 108–119 (2017).
Jensen, M. C. & Riddell, S. R. Designing chimeric antigen receptors to effectively and safely target tumors. Curr. Opin. Immunol. 33, 9–15 (2015).
Guest, R. D. et al. The role of extracellular spacer regions in the optimal design of chimeric immune receptors: evaluation of four different scFvs and antigens. J. Immunother. 28, 203–211 (2005).
Li, J. et al. Membrane-proximal epitope facilitates efficient t cell synapse formation by anti-FcRH5/CD3 and is a requirement for myeloma cell killing. Cancer Cell 31, 383–395 (2017).
Gradisar, H. et al. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat. Chem. Biol. 9, 362–366 (2013).
Goodman, R. P. et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310, 1661–1665 (2005).
Dustin, M. L., Starr, T., Varma, R. & Thomas, V. K. Supported planar bilayers for study of the immunological synapse. Curr. Protoc. Immunol. 18, 11–35 (2007).
Cai, H. et al. Spatial control of biological ligands on surfaces applied to T cell activation. Methods Mol. Biol. 1584, 307–331 (2017).
Vasiliver-Shamis, G., Cho, M. W., Hioe, C. E. & Dustin, M. L. Human immunodeficiency virus type 1 envelope gp120-induced partial T-cell receptor signaling creates an F-actin-depleted zone in the virological synapse. J. Virol. 83, 11341–11355 (2009).
Acknowledgements
The authors thank S. Curado for coordination and S. Davis for insightful comments. This work was supported primarily by the National Science Foundation under award no. CMMI-1300590, by the National Institutes of Health Common Fund Nanomedicine programme, grants PN2 EY016586, R37 AI043542 and P01 A1080192; and Wellcome Trust and Kennedy Trust for Rheumatology Research PRF 100262Z/12/Z. The Columbia Nano Initiative provided cleanroom and processing facilities. We thank M. Cammer of NYULMC OCR for microscopy and analysis support.
Author information
Authors and Affiliations
Contributions
H.C. carried out nanofabrication. H.C. and D.D. contributed equally to development of the bilayer backfill methodology. H.C. and J.M. contributed equally to the data acquisition and analysis. V.M. provided purified ICAM1. S.J.W., M.L.D. and M.P.S. designed the experiments. H.C., J.M., M.L.D. and S.J.W. interpreted data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–13, Supplementary Tables 1–2, Supplementary References.
Rights and permissions
About this article
Cite this article
Cai, H., Muller, J., Depoil, D. et al. Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering. Nature Nanotech 13, 610–617 (2018). https://doi.org/10.1038/s41565-018-0113-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-018-0113-3
This article is cited by
-
Membrane-anchored DNA nanojunctions enable closer antigen-presenting cell–T-cell contact in elevated T-cell receptor triggering
Nature Nanotechnology (2023)
-
Harnessing Biomaterials for Immunomodulatory-Driven Tissue Engineering
Regenerative Engineering and Translational Medicine (2023)
-
Precise regulation of intermembrane spacing with DNA nanojunctions for enhanced T-cell receptor triggering
Science China Chemistry (2023)
-
The interplay between membrane topology and mechanical forces in regulating T cell receptor activity
Communications Biology (2022)
-
Ligand functionalization of titanium nanopattern enables the analysis of cell–ligand interactions by super-resolution microscopy
Nature Protocols (2022)