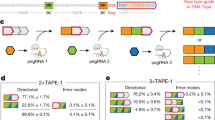
Abstract
Fundamental computer science concepts have inspired novel information-processing molecular systems in test tubes1,2,3,4,5,6,7,8,9,10,11,12,13 and genetically encoded circuits in live cells14,15,16,17,18,19,20,21. Recent research has shown that digital information storage in DNA, implemented using deep sequencing and conventional software, can approach the maximum Shannon information capacity22 of two bits per nucleotide23. In nature, DNA is used to store genetic programs, but the information content of the encoding rarely approaches this maximum24. We hypothesize that the biological function of a genetic program can be preserved while reducing the length of its DNA encoding and increasing the information content per nucleotide. Here we support this hypothesis by describing an experimental procedure for compressing a genetic program and its subsequent autonomous decompression and execution in human cells. As a test-bed we choose an RNAi cell classifier circuit25 that comprises redundant DNA sequences and is therefore amenable for compression, as are many other complex gene circuits15,18,26,27,28. In one example, we implement a compressed encoding of a ten-gene four-input AND gate circuit using only four genetic constructs. The compression principles applied to gene circuits can enable fitting complex genetic programs into DNA delivery vehicles with limited cargo capacity, and storing compressed and biologically inert programs in vivo for on-demand activation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Adleman, L. M. Molecular computation of solutions to combinatorial problems. Science 266, 1021–1024 (1994).
Mao, C. D., LaBean, T. H., Reif, J. H. & Seeman, N. C. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature 407, 493–496 (2000).
Benenson, Y. et al. Programmable and autonomous computing machine made of biomolecules. Nature 414, 430–434 (2001).
Qian, L., Winfree, E. & Bruck, J. Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011).
Liu, Q. et al. DNA computing on surfaces. Nature 403, 175–179 (2000).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Phillips, A. & Cardelli, L. A programming language for composable DNA circuits. J. Royal Soc. Interface 6, S419–S436 (2009).
Gehani, A., LaBean, T. & Reif, J. DNA-based cryptography. Lecture Notes Comp. Sci. 2950, 167–188 (2003).
Chen, Y.-J. et al. Programmable chemical controllers made from DNA. Nat. Nanotech. 8, 755–762 (2013).
Church, G. M., Gao, Y. & Kosuri, S. Next-generation digital information storage in DNA. Science 337, 1628 (2012).
Padirac, A., Fujii, T. & Rondelez, Y. Bottom-up construction of in vitro switchable memories. Proc. Natl Acad. Sci. USA 109, E3212–E3220 (2012).
Weitz, M. et al. Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nat. Chem. 6, 295–302 (2014).
Stojanovic, M. N. & Stefanovic, D. A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 21, 1069–1074 (2003).
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Rinaudo, K. et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 25, 795–801 (2007).
Regot, S. et al. Distributed biological computation with multicellular engineered networks. Nature 469, 207–211 (2011).
Bonnet, J., Yin, P., Ortiz, M. E., Subsoontorn, P. & Endy, D. Amplifying genetic logic gates. Science 340, 599–603 (2013).
Daniel, R., Rubens, J. R., Sarpeshkar, R. & Lu, T. K. Synthetic analog computation in living cells. Nature 497, 619–623 (2013).
Nielsen, A. et al. Genetic circuit design automation. Science 352, aac7341 (2016).
Green, A. A., Silver, Pamela, A., Collins, James, J. & Yin, P. Toehold switches: de-novo-designed regulators of gene expression. Cell 159, 925–939 (2014).
Anderson, J. C., Voigt, C. A. & Arkin, A. P. Environmental signal integration by a modular and gate. Mol. Syst. Biol. 3, 133 (2007).
Shannon, C. E. A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 (1948). 623–656.
Erlich, Y. & Zielinski, D. DNA fountain enables a robust and efficient storage architecture. Science 355, 950–954 (2017).
Kuruppu, S., Puglisi, S. & Zobel, J. Optimized relative Lempel–Ziv compression of genomes. Proc. 34th Australas. Comp. Sci. Conf. 91–98 (2011).
Xie, Z., Wroblewska, L., Prochazka, L., Weiss, R. & Benenson, Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311 (2011).
Xie, M. et al. β-cell-mimetic designer cells provide closed-loop glycemic control. Science 354, 1296–1301 (2016).
Prochazka, L., Angelici, B., Haefliger, B. & Benenson, Y. Highly modular bow-tie gene circuits with programmable dynamic behaviour. Nat. Commun. 5, 4729 (2014).
Schreiber, J., Arter, M., Lapique, N., Haefliger, B. & Benenson, Y. Model-guided combinatorial optimization of complex synthetic gene networks. Mol. Syst. Biol. 12, 899 (2016).
Heagerty, P., Lumley, T. & Pepe, M. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 56, 337–344 (2000).
Lapique, N. & Benenson, Y. Digital switching in a biosensor circuit via programmable timing of gene availability. Nat. Chem. Biol. 10, 1020–1027 (2014).
Bhatia, S., LaBoda, C., Yanez, V., Haddock-Angelli, T. & Densmore, D. Permutation machines. ACS Syn. Biol. 5, 827–834 (2016).
Ham, T. S., Lee, S. K., Keasling, J. D. & Arkin, A. P. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE 3, e2815 (2008).
Haefliger, B., Prochazka, L., Angelici, B. & Benenson, Y. Precision multidimensional assay for high-throughput microRNA drug discovery.Nat. Commun. 7, 10709 (2016).
Acknowledgements
The research was funded by the National Institutes of Health award 5R01CA155320 and by ETH Zürich. We thank B. Angelici for discussions and E. Shapiro for commenting on the manuscript.
Author information
Authors and Affiliations
Contributions
N.L. conceived research, performed experiments, analysed data, and wrote the paper. Y.B. conceived research, analysed data, supervised the project, and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The original miRNA circuit technology is protected by patents awarded to Y.B. and co-inventors (US patent no. 9458509). The output delay technology is pending, with N.L. and Y.B. listed as co-inventors.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lapique, N., Benenson, Y. Genetic programs can be compressed and autonomously decompressed in live cells. Nature Nanotech 13, 309–315 (2018). https://doi.org/10.1038/s41565-017-0004-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-017-0004-z
This article is cited by
-
Engineering intelligent chassis cells via recombinase-based MEMORY circuits
Nature Communications (2024)
-
Biological factors in the synthetic construction of overlapping genes
BMC Genomics (2021)
-
Engineering calcium signaling of astrocytes for neural–molecular computing logic gates
Scientific Reports (2021)
-
Large-scale DNA-based phenotypic recording and deep learning enable highly accurate sequence-function mapping
Nature Communications (2020)
-
Encryption and steganography of synthetic gene circuits
Nature Communications (2018)