Abstract
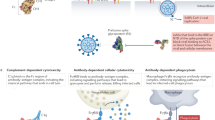
Antibodies serve as critical barriers to viral infection. Humoral immunity to a virus is achieved through the dual role of antibodies in communicating the presence of invading pathogens in infected cells to effector cells, and in interfering with processes essential to the viral life cycle (chiefly entry into the host cell). For individuals that successfully control infection, virus-elicited antibodies can provide lifelong surveillance and protection from future insults. One approach to understand the nature of a successful immune response has been to utilize structural biology to uncover the molecular details of antibodies derived from vaccines or natural infection and how they interact with their cognate microbial antigens. The ability to isolate antigen-specific B-cells and rapidly solve structures of functional, monoclonal antibodies in complex with viral glycoprotein surface antigens has greatly expanded our knowledge of the sites of vulnerability on viruses. In this Review, we compare the adaptive humoral immune responses to human immunodeficiency virus (HIV), influenza and filoviruses, with a particular focus on neutralizing antibodies. The pathogenesis of each of these viruses is quite different, providing an opportunity for comparison of immune responses: HIV causes a persistent, chronic infection; influenza, an acute infection with multiple exposures during a lifetime and annual vaccination; filoviruses, a virulent, acute infection. Neutralizing antibodies that develop under these different constraints are therefore sentinels that can provide insight into the underlying humoral immune responses, as well as important lessons to guide future development of vaccines and immunotherapeutics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
White, J. M. & Whittaker, G. R. Fusion of enveloped viruses in endosomes. Traffic 17, 593–614 (2016).
Harrison, S. C. Viral membrane fusion. Virology 479–480, 498–507 (2015).
Harris, L. J., Larson, S. B., Hasel, K. W. & McPherson, A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 36, 1581–1597 (1997).
Harris, L. J., Skaletsky, E. & McPherson, A. Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 275, 861–872 (1998).
Saphire, E. O. et al. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293, 1155–1159 (2001).
Scapin, G. et al. Structure of full-length human anti-PD1 therapeutic IgG4 antibody pembrolizumab. Nat. Struct. Mol. Biol. 22, 953–958 (2015).
Stanfield, R. L. & Wilson, I. A. Antibody Structure. Microbiol. Spectr. 2, AID-0012-2013 (2014).
Wilson, I. A. & Stanfield, R. L. Antibody-antigen interactions: new structures and new conformational changes. Curr. Opin. Struct. Biol. 4, 857–867 (1994).
Bullough, P. A., Hughson, F. M., Skehel, J. J. & Wiley, D. C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 (1994).
Wilson, I. A., Skehel, J. J. & Wiley, D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289, 366–373 (1981).
Julien, J. P. et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 (2013).
Lyumkis, D. et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 (2013).
Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997).
Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997).
Lee, J. E. et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182 (2008).
Weissenhorn, W., Carfi, A., Lee, K. H., Skehel, J. J. & Wiley, D. C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2, 605–616 (1998).
Cook, J. D. & Lee, J. E. The secret life of viral entry glycoproteins: moonlighting in immune evasion. PLoS Pathog. 9, e1003258 (2013).
Tate, M. D. et al. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6, 1294–1316 (2014).
Wu, N. C. & Wilson, I. A. A Perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J. Mol. Biol. 429, 2694–2709 (2017).
Crispin, M., Ward, A. B. & Wilson, I. A. Structure and immune recognition of the HIV glycan shield. Ann. Rev. Biophys. 47, 499–523 (2018).
Cao, L. et al. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat. Commun. 8, 14954 (2017).
Lee, J. H., de Val, N., Lyumkis, D. & Ward, A. B. Model building and refinement of a natively glycosylated HIV-1 Env protein by high-resolution cryoelectron microscopy. Structure 23, 1943–1951 (2015).
Ekiert, D. C. et al. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 (2009).
Flyak, A. I. et al. Mechanism of human antibody-mediated neutralization of Marburg virus. Cell 160, 893–903 (2015).
Julien, J. P. et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl Acad. Sci. USA 110, 4351–4356 (2013).
Kong, L. et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 20, 796–803 (2013).
Murin, C. D. et al. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J. Virol. 88, 10177–10188 (2014).
Williams, K. L. et al. Superinfection drives HIV neutralizing antibody responses from several B cell lineages that contribute to a polyclonal repertoire. Cell Rep. 23, 682–691 (2018).
Bianchi, M. et al. Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity 49, 288–300 (2018).
Ozorowski, G. et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547, 360–363 (2017).
Georgiev, I. S., Gordon Joyce, M., Zhou, T. & Kwong, P. D. Elicitation of HIV-1-neutralizing antibodies against the CD4-binding site. Curr. Opin. HIV AIDS 8, 382–392 (2013).
Zhou, T. et al. Structural Repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161, 1280–1292 (2015).
Xu, R. et al. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat. Struct. Mol. Biol. 20, 363–370 (2013).
Garcia-Sastre, A. Influenza virus receptor specificity: disease and transmission. Am. J. Pathol. 176, 1584–1585 (2010).
Stevens, J. et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–410 (2006).
Wu, N. C. et al. A complex epistatic network limits the mutational reversibility in the influenza hemagglutinin receptor-binding site. Nat. Commun. 9, 1264 (2018).
Wu, N. C. et al. Diversity of functionally permissive sequences in the receptor-binding site of influenza hemagglutinin. Cell Host Microbe 22, 247–248 (2017).
Lee, P. S. et al. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 5, 3614 (2014).
Wu, N. C. et al. In vitro evolution of an influenza broadly neutralizing antibody is modulated by hemagglutinin receptor specificity. Nat. Commun. 8, 15371 (2017).
Julien, J. P., Lee, P. S. & Wilson, I. A. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol. Rev. 250, 180–198 (2012).
Laursen, N. S. & Wilson, I. A. Broadly neutralizing antibodies against influenza viruses. Antivir. Res. 98, 476–483 (2013).
Lee, P. S. & Wilson, I. A. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr. Top. Microbiol. 386, 323–341 (2015).
Bornholdt, Z. A. et al. Host-primed Ebola virus GP exposes a hydrophobic NPC1 receptor-binding pocket, revealing a target for broadly neutralizing antibodies. mBio 7, e02154–15 (2016).
Chandran, K., Sullivan, N. J., Felbor, U., Whelan, S. P. & Cunningham, J. M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645 (2005).
Miller, E. H. et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 31, 1947–1960 (2012).
Gnirss, K. et al. Cathepsins B and L activate Ebola but not Marburg virus glycoproteins for efficient entry into cell lines and macrophages independent of TMPRSS2 expression. Virology 424, 3–10 (2012).
King, L. B. et al. The Marburgvirus-neutralizing human monoclonal antibody MR191 targets a conserved site to block virus receptor binding. Cell Host Microbe 23, 101–109 (2018).
Hashiguchi, T. et al. Structural basis for Marburg virus neutralization by a cross-reactive human antibody. Cell 160, 904–912 (2015).
Wang, H. et al. Ebola viral glycoprotein bound to its endosomal receptor niemann-pick C1. Cell 164, 258–268 (2016).
Lin, Y. P. et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc. Natl Acad. Sci. USA 109, 21474–21479 (2012).
Ekiert, D. C. et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532 (2012).
Lee, P. S. et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc. Natl Acad. Sci. USA 109, 17040–17045 (2012).
Whittle, J. R. et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl Acad. Sci. USA 108, 14216–14221 (2011).
Schmidt, A. G. et al. Viral receptor-binding site antibodies with diverse germline origins. Cell 161, 1026–1034 (2015).
Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Garrett, T. P., Wang, J., Yan, Y., Liu, J. & Harrison, S. C. Refinement and analysis of the structure of the first two domains of human CD4. J. Mol. Biol. 234, 763–778 (1993).
Wu, X. et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010).
Zhou, T. et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 (2010).
Zhou, T. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 (2007).
Bale, S. et al. Structural basis for differential neutralization of ebolaviruses. Viruses 4, 447–470 (2012).
Dias, J. M. et al. A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol. 18, 1424–1427 (2011).
Lee, J. E. & Saphire, E. O. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 4, 621–635 (2009).
Crowe, J. E. Jr. Principles of broad and potent antiviral human antibodies: insights for vaccine design. Cell Host Microbe 22, 193–206 (2017).
Hashem, A. M. et al. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem. Bioph. Res. Co. 403, 247–251 (2010).
Murin, C. D., Bruhn, J. F., Bornholdt, Z. A., Copps, J., Stanfield, R. & Ward, A. B. Structural basis of pan-ebolavirus neutralization by an antibody targeting the glycoprotein fusion loop. Cell Rep. 24, 2723–2732 (2018).
van Gils, M. J. et al. An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat. Microbiol. 2, 16199 (2016).
Kong, R. et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833 (2016).
Xu, K. et al. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 24, 857–867 (2018).
Kallewaard, N. L. et al. Structure and function analysis of an antibody recognizing all Influenza A subtypes. Cell 166, 596–608 (2016).
Prabhu, N. et al. Monoclonal antibodies against the fusion peptide of hemagglutinin protect mice from lethal influenza A virus H5N1 infection. J. Virol. 83, 2553–2562 (2009).
Zhao, X. et al. Immunization-elicited broadly protective antibody reveals Ebolavirus fusion loop as a site of vulnerability. Cell 169, 891–904 (2017).
Furuyama, W. et al. Discovery of an antibody for pan-ebolavirus therapy. Sci. Rep. 6, 20514 (2016).
Wec, A. Z. et al. Antibodies from a human survivor define sites of vulnerability for broad protection against Ebolaviruses. Cell 169, 878–890 (2017).
Zhu, P. et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl Acad. Sci. USA 100, 15812–15817 (2003).
Zhu, P. et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 (2006).
Lee, J. H., Ozorowski, G. & Ward, A. B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048 (2016).
Huang, J. et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012).
Cardoso, R. M. et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22, 163–173 (2005).
Flyak, A. I. et al. Broadly neutralizing antibodies from human survivors target a conserved site in the Ebola virus glycoprotein HR2-MPER region. Nat. Microbiol. 3, 670–677 (2018).
Krammer, F. & Palese, P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol. 3, 521–530 (2013).
Dreyfus, C., Ekiert, D. C. & Wilson, I. A. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J. Virol. 87, 7149–7154 (2013).
Khanna, M., Sharma, S., Kumar, B. & Rajput, R. Protective immunity based on the conserved hemagglutinin stalk domain and its prospects for universal influenza vaccine development. Biomed. Res. Int. 2014, 546274 (2014).
Erbelding, E. J. et al. A universal Influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 218, 347–354 (2018).
Impagliazzo, A. et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306 (2015).
Krammer, F., Garcia-Sastre, A. & Palese, P. Is it possible to develop a “universal” influenza virus vaccine? Potential target antigens and critical aspects for a universal influenza vaccine. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a028845 (2018).
Krammer, F., Pica, N., Hai, R., Margine, I. & Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 87, 6542–6550 (2013).
Nachbagauer, R. et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines 2, 26 (2017).
Yassine, H. M. et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 21, 1065–1070 (2015).
Irimia, A. et al. Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design. PLoS Pathog. 13, e1006212 (2017).
Irimia, A., Sarkar, A., Stanfield, R. L. & Wilson, I. A. Crystallographic identification of lipid as an integral component of the epitope of HIV broadly neutralizing antibody 4E10. Immunity 44, 21–31 (2016).
Friesen, R. H. et al. A common solution to group 2 influenza virus neutralization. Proc. Natl Acad. Sci. USA 111, 445–450 (2014).
Burton, D. R. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2, 706–713 (2002).
Burton, D. R. & Mascola, J. R. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 16, 571–576 (2015).
Burton, D. R., Poignard, P., Stanfield, R. L. & Wilson, I. A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337, 183–186 (2012).
Burton, D. R., Stanfield, R. L. & Wilson, I. A. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl Acad. Sci. USA 102, 14943–14948 (2005).
Caskey, M., Klein, F. & Nussenzweig, M. C. Broadly neutralizing antibodies for HIV-1 prevention or immunotherapy. N. Engl. J. Med. 375, 2019–2021 (2016).
Mouquet, H. et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl Acad. Sci. USA 109, E3268–3277 (2012).
McLellan, J. S. et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 (2011).
Julien, J. P. et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 9, e1003342 (2013).
Pejchal, R. et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 (2011).
Calarese, D. A. et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300, 2065–2071 (2003).
Stanfield, R. L., De Castro, C., Marzaioli, A. M., Wilson, I. A. & Pantophlet, R. Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose. Glycobiology 25, 412–419 (2015).
Kwong, P. D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002).
Lee, J. H. et al. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic beta-hairpin structure. Immunity 46, 690–702 (2017).
Cale, E. M. et al. Virus-like particles identify an HIV V1V2 apex-binding neutralizing antibody that lacks a protruding loop. Immunity 46, 777–791 (2017).
Wang, H. et al. Asymmetric recognition of HIV-1 envelope trimer by V1V2 loop-targeting antibodies. eLife 6, e27389 (2017).
Kashyap, A. K. et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl Acad. Sci. USA 105, 5986–5991 (2008).
Lang, S. et al. Antibody 27F3 broadly targets influenza A group 1 and 2 hemagglutinins through a further variation in VH1–69 antibody orientation on the HA stem. Cell Rep. 20, 2935–2943 (2017).
Sui, J. et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16, 265–273 (2009).
Throsby, M. et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 3, e3942 (2008).
Lerner, R. A. Rare antibodies from combinatorial libraries suggests an S. O. S. component of the human immunological repertoire. Mol. Biosyst. 7, 1004–1012 (2011).
Lerner, R. A. Combinatorial antibody libraries: new advances, new immunological insights. Nat. Rev. Immunol. 16, 498–508 (2016).
Pallesen, J. et al. Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nat. Microbiol. 1, 16128 (2016).
de La Vega, M. A., Wong, G., Kobinger, G. P. & Qiu, X. The multiple roles of sGP in Ebola pathogenesis. Viral Immunol. 28, 3–9 (2015).
Bornholdt, Z. A. et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 351, 1078–1083 (2016).
Flyak, A. I. et al. Cross-reactive and potent neutralizing antibody responses in human survivors of natural ebolavirus infection. Cell 164, 392–405 (2016).
Wilson, J. A. et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science 287, 1664–1666 (2000).
Howell, K. A. et al. Cooperativity enables non-neutralizing antibodies to neutralize ebolavirus. Cell Rep. 19, 413–424 (2017).
Kwong, P. D. & Mascola, J. R. HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871 (2018).
Flynn, N. M. et al. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191, 654–665 (2005).
Pitisuttithum, P. et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 194, 1661–1671 (2006).
Dolin, R. et al. The safety and immunogenicity of a human immunodeficiency virus type 1 (HIV-1) recombinant gp160 candidate vaccine in humans. NIAID AIDS Vaccine Clinical Trials Network. Ann. Intern. Med. 114, 119–127 (1991).
Cooney, E. L. et al. Safety of and immunological response to a recombinant vaccinia virus vaccine expressing HIV envelope glycoprotein. Lancet 337, 567–572 (1991).
Burton, D. R. et al. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5, 233–236 (2004).
Gonzalez, N. et al. Characterization of broadly neutralizing antibody responses to HIV-1 in a cohort of long term non-progressors. PLoS ONE 13, e0193773 (2018).
Gray, E. S. et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85, 4828–4840 (2011).
Kelsoe, G. & Haynes, B. F. Host controls of HIV broadly neutralizing antibody development. Immunol. Rev. 275, 79–88 (2017).
Burton, D. R. et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 (1994).
Muster, T. et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67, 6642–6647 (1993).
Trkola, A. et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70, 1100–1108 (1996).
Buchacher, A. et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrov. 10, 359–369 (1994).
Binley, J. M. et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74, 627–643 (2000).
Sanders, R. W. et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76, 8875–8889 (2002).
Sanders, R. W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013).
Sanders, R. W. & Moore, J. P. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev. 275, 161–182 (2017).
Sanders, R. W., Wilson, I. A. & Moore, J. P. HIV’s Achilles’ heel. Sci. Am. 315, 50–55 (2016).
Torrents de la Pena, A. et al. Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 20, 1805–1817 (2017).
Voss, J. E. et al. Elicitation of neutralizing antibodies targeting the V2 apex of the HIV envelope trimer in a wild-type animal model. Cell Rep. 21, 222–235 (2017).
de Taeye, S. W. et al. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163, 1702–1715 (2015).
Kulp, D. W. & Schief, W. R. Advances in structure-based vaccine design. Curr. Opin. Virol. 3, 322–331 (2013).
Kulp, D. W. et al. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat. Commun. 8, 1655 (2017).
Correia, B. E. et al. Proof of principle for epitope-focused vaccine design. Nature 507, 201–206 (2014).
Jardine, J. et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013).
Jardine, J. G. et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351, 1458–1463 (2016).
Jardine, J. G. et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161 (2015).
Correia, B. E. et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 18, 1116–1126 (2010).
McLellan, J. S. et al. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J. Mol. Biol. 409, 853–866 (2011).
Ofek, G. et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl Acad. Sci. USA 107, 17880–17887 (2010).
de Taeye, S. W., Moore, J. P. & Sanders, R. W. HIV-1 Envelope trimer design and immunization strategies to induce broadly neutralizing antibodies. Trends Immunol. 37, 221–232 (2016).
Sanders, R. W. et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223 (2015).
Pauthner, M. et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46, 1073–1088 (2017).
Krammer, F. et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J. Virol. 88, 3432–3442 (2014).
Krammer, F. et al. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J. Virol. 88, 2340–2343 (2014).
Mallajosyula, V. V. et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc. Natl Acad. Sci. USA 111, E2514–E2523 (2014).
Wohlbold, T. J. et al. Vaccination with soluble headless hemagglutinin protects mice from challenge with divergent influenza viruses. Vaccine 33, 3314–3321 (2015).
Hai, R. et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J. Virol. 86, 5774–5781 (2012).
Goo, L., Chohan, V., Nduati, R. & Overbaugh, J. Early development of broadly neutralizing antibodies in HIV-1-infected infants. Nat. Med. 20, 655–658 (2014).
van den Kerkhof, T. L. et al. Early development of broadly reactive HIV-1 neutralizing activity in elite neutralizers. AIDS 28, 1237–1240 (2014).
Sok, D. et al. Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557–1560 (2016).
Havenar-Daughton, C. et al. The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl Med. 10, eaat0381 (2018).
Havenar-Daughton, C., Abbott, R. K., Schief, W. R. & Crotty, S. When designing vaccines, consider the starting material: the human B cell repertoire. Curr. Opin. Immunol. 53, 209–216 (2018).
Arnaout, R. et al. High-resolution description of antibody heavy-chain repertoires in humans. PLoS ONE 6, e22365 (2011).
DeKosky, B. J. et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat. Med. 21, 86–91 (2015).
Abbott, R. K. et al. Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity 48, 133–146 (2018).
Havenar-Daughton, C. et al. Direct probing of germinal center responses reveals immunological features and bottlenecks for neutralizing antibody responses to HIV Env trimer. Cell Rep. 17, 2195–2209 (2016).
Doria-Rose, N. A. et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62 (2014).
Bonsignori, M. et al. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 165, 449–463 (2016).
Rantalainen, K. et al. Co-evolution of HIV envelope and apex-targeting neutralizing antibody lineage provides benchmarks for vaccine design. Cell Rep. 23, 3249–3261 (2018).
Landais, E. et al. Broadly neutralizing antibody responses in a large longitudinal Sub-Saharan HIV primary infection cohort. PLoS Pathog. 12, e1005369 (2016).
Landais, E. et al. HIV Envelope glycoform heterogeneity and localized diversity govern the Initiation and maturation of a V2 apex broadly neutralizing antibody lineage. Immunity 47, 990–1003 (2017).
Doria-Rose, N. A. et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. 90, 76–91 (2016).
Dosenovic, P. et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell 161, 1505–1515 (2015).
Escolano, A. et al. Sequential immunization elicits broadly neutralizing anti-HIV-1 antibodies in Ig knockin mice. Cell 166, 1445–1458 (2016).
Lee, E. C. et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol. 32, 356–363 (2014).
Acharya, P. et al. CD4-binding-site recognition by VH1–46 germline-derived HIV-1 neutralizers. AIDS Res. Hum. Retrov. 30, A120–A121 (2014).
Gorny, M. K. et al. Preferential use of the VH5–51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol. Immunol. 46, 917–926 (2009).
Huang, C. C. et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl Acad. Sci. USA 101, 2706–2711 (2004).
Baize, S. et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 371, 1418–1425 (2014).
Oswald, W. B. et al. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 3, e9 (2007).
Qiu, X. et al. Sustained protection against Ebola virus infection following treatment of infected nonhuman primates with ZMAb. Sci. Rep. 3, 3365 (2013).
Pettitt, J. et al. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci. Transl Med. 5, 199ra113 (2013).
Murin, C. D. et al. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc. Natl Acad. Sci. USA 111, 17182–17187 (2014).
Qiu, X. et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53 (2014).
Group, P. I. W. et al. A randomized, controlled trial of ZMapp for ebola virus infection. N. Engl. J. Med. 375, 1448–1456 (2016).
Henao-Restrepo, A. M. et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ca Suffit!). Lancet 389, 505–518 (2017).
Rimoin, A. W. et al. Ebola virus neutralizing antibodies detectable in survivors of the Yambuku, Zaire outbreak 40 years after infection. J. Infect. Dis. 217, 223–231 (2018).
Pascal, K. E. et al. Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in non-human primates. J. Infect. Dis. 218, 612–626 (2018).
Misasi, J. et al. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J. Virol. 86, 3284–3292 (2012).
Saphire, E. O. et al. Systematic analysis of monoclonal antibodies against Ebola virus GP defines features that contribute to protection. Cell 174, 938–952 (2018).
Murphy, A. J. et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. P. Natl Acad. Sci. USA 111, 5153–5158 (2014).
Beniac, D. R. et al. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS ONE 7, e29608 (2012).
Harris, A. et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl Acad. Sci. USA 103, 19123–19127 (2006).
Liu, J., Bartesaghi, A., Borgnia, M. J., Sapiro, G. & Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 (2008).
Liu, J., Wright, E. R. & Winkler, H. 3D visualization of HIV virions by cryoelectron tomography. Method Enzymol. 483, 267–290 (2010).
Klein, J. S. & Bjorkman, P. J. Few and far between: How HIV may be evading antibody avidity. PLoS Pathog. 6, e1000908 (2010).
Galimidi, R. P. et al. Intra-spike crosslinking overcomes antibody evasion by HIV-1. Cell 160, 433–446 (2015).
Walker, L. M. et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009).
Scheid, J. F. et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640 (2009).
Simek, M. D. et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83, 7337–7348 (2009).
Walker, L. M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011).
Cirelli, K. M. & Crotty, S. Germinal center enhancement by extended antigen availability. Curr. Opin. Immunol. 47, 64–69 (2017).
Crotty, S. Raging evolution of a B cell response to a viral infection. Nat. Rev. Immunol. 18, 79 (2018).
Horwitz, J. A. et al. Non-neutralizing antibodies alter the course of HIV-1 infection in vivo. Cell 170, 637–648 (2017).
Mayr, L., Su, B. & Moog, C. Role of non-neutralizing antibodies in vaccines and/or HIV infected individuals. Curr. Opin. HIV AIDS 12, 209–215 (2017).
Forthal, D., Hope, T. J. & Alter, G. New paradigms for functional HIV-specific non-neutralizing antibodies. Curr. Opin. HIV AIDS 8, 393–401 (2013).
Kajihara, M. et al. Inhibition of Marburg virus budding by non-neutralizing antibodies to the envelope glycoprotein. J. Virol. 86, 13467–13474 (2012).
Bournazos, S. et al. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158, 1243–1253 (2014).
Bournazos, S. & Ravetch, J. V. Anti-retroviral antibody FcγR-mediated effector functions. Immunol. Rev. 275, 285–295 (2017).
Bournazos, S. & Ravetch, J. V. Fcγ receptor function and the design of vaccination strategies. Immunity 47, 224–233 (2017).
Nimmerjahn, F. & Ravetch, J. V. Fcγ receptors: old friends and new family members. Immunity 24, 19–28 (2006).
Nimmerjahn, F. & Ravetch, J. V. Fc-receptors as regulators of immunity. Adv. Immunol. 96, 179–204 (2007).
Nimmerjahn, F. & Ravetch, J. V. Fcγ receptors as regulators of immune responses. Nat. Rev. Immunol. 8, 34–47 (2008).
Pelegrin, M., Naranjo-Gomez, M. & Piechaczyk, M. Antiviral monoclonal antibodies: can they be more than simple neutralizing agents? Trends Microbiol. 23, 653–665 (2015).
Acknowledgements
This work was supported by the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (grant no. NIAID UM1AI100663), the Viral Hemorrhagic Fever Immunotherapeutic Consortium (grant no. U19 AI109762), NIH (grant no. R56 AI127371), the Bill & Melinda Gates Collaboration for AIDS Vaccine Discovery (grant no. OPP1084519) and the International AIDS Vaccine Initiative.
Author information
Authors and Affiliations
Contributions
C. D. M. wrote the initial draft; C. D. M., I. A. W. and A. B. W. wrote, reviewed and edited the Review.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Murin, C.D., Wilson, I.A. & Ward, A.B. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat Microbiol 4, 734–747 (2019). https://doi.org/10.1038/s41564-019-0392-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0392-y
This article is cited by
-
Computational design and evaluation of mRNA- and protein-based conjugate vaccines for influenza A and SARS-CoV-2 viruses
Journal of Genetic Engineering and Biotechnology (2023)
-
Antigen epitopes of animal coronaviruses: a mini-review
Animal Diseases (2023)
-
Vaccination with a replication-defective cytomegalovirus vaccine elicits a glycoprotein B-specific monoclonal antibody repertoire distinct from natural infection
npj Vaccines (2023)
-
Antiviral neutralizing antibodies: from in vitro to in vivo activity
Nature Reviews Immunology (2023)
-
The Landscape of Neutralizing Monoclonal Antibodies (nAbs) for Treatment and Prevention of COVID-19
Journal of Pharmaceutical Innovation (2023)