Abstract
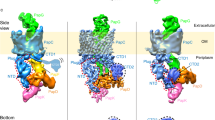
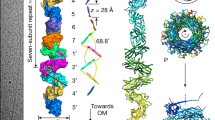
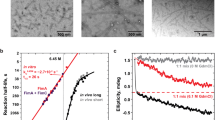
Chaperone–usher pathway pili are extracellular proteinaceous fibres ubiquitously found on Gram-negative bacteria, and mediate host–pathogen interactions and biofilm formation critical in pathogenesis in numerous human diseases1. During pilus assembly, an outer membrane macromolecular machine called the usher catalyses pilus biogenesis from the individual subunits that are delivered as chaperone–subunit complexes in the periplasm. The usher orchestrates pilus assembly using all five functional domains: a 24-stranded transmembrane β-barrel translocation domain, a β-sandwich plug domain, an amino-terminal periplasmic domain and two carboxy-terminal periplasmic domains (CTD1 and CTD2)2,3,4,5,6. Despite extensive structural and functional characterization, the mechanism by which the usher is activated to initiate pilus biogenesis is unknown. Here, we present the crystal structure of the full-length PapC usher from Escherichia coli in complex with its cognate PapDG chaperone–subunit complex in a pre-activation state, elucidating molecular details of how the usher is specifically engaged by allosteric interactions with its substrate preceding activation and how the usher facilitates the transfer of subunits from the amino-terminal periplasmic domain to the CTDs during pilus assembly. This work elucidates the intricate workings of a molecular machine that catalyses chaperone–usher pathway pilus assembly and opens the door for the development of potent inhibitors to block pilus biogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon request. Atomic coordinates and structure factors for the reported crystal structure have been deposited into the Protein Data Bank under accession code 6CD2.
References
Nuccio, S. P. & Baumler, A. J. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71, 551–575 (2007).
Ford, B. et al. Structural homology between the C-terminal domain of the PapC usher and its plug. J. Bacteriol. 192, 1824–1831 (2010).
Nishiyama, M. et al. Structural basis of chaperone–subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 24, 2075–2086 (2005).
Phan, G. et al. Crystal structure of the FimD usher bound to its cognate FimC–FimH substrate. Nature 474, 49–53 (2011).
Remaut, H. et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133, 640–652 (2008).
Thanassi, D. G., Stathopoulos, C., Dodson, K., Geiger, D. & Hultgren, S. J. Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J. Bacteriol. 184, 6260–6269 (2002).
Foxman, B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113, 5S–13S (2002).
Foxman, B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis. Mon. 49, 53–70 (2003).
Wurpel, D. J., Beatson, S. A., Totsika, M., Petty, N. K. & Schembri, M. A. Chaperone–usher fimbriae of Escherichia coli. PLoS ONE 8, e52835 (2013).
Ronald, A. The etiology of urinary tract infection: traditional and emerging pathogens. Dis. Mon. 49, 71–82 (2003).
Ronald, A. R. et al. Urinary tract infection in adults: research priorities and strategies. Int. J. Antimicrob. Agents 17, 343–348 (2001).
Melican, K. et al. Uropathogenic Escherichia coli P and type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 7, e1001298 (2011).
Roberts, J. A. et al. The Gal(α1-4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl Acad. Sci. USA 91, 11889–11893 (1994).
Abraham, S. N., Sun, D., Dale, J. B. & Beachey, E. H. Conservation of the d-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature 336, 682–684 (1988).
Lindberg, F., Tennent, J. M., Hultgren, S. J., Lund, B. & Normark, S. PapD, a periplasmic transport protein in P-pilus biogenesis. J. Bacteriol. 171, 6052–6058 (1989).
Hultgren, S. J. et al. The PapG adhesin of uropathogenic Escherichia coli contains separate regions for receptor binding and for the incorporation into the pilus. Proc. Natl Acad. Sci. USA 86, 4357–4361 (1989).
Choudhury, D. et al. X-ray structure of the FimC–FimH chaperone–adhesin complex from uropathogenic Escherichia coli. Science 285, 1061–1066 (1999).
Sauer, F. G. et al. Structural basis of chaperone function and pilus biogenesis. Science 285, 1058–1061 (1999).
Zavialov, A. V. et al. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113, 587–596 (2003).
Remaut, H. et al. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol. Cell 22, 831–842 (2006).
Sauer, F. G., Pinkner, J. S., Waksman, G. & Hultgren, S. J. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111, 543–551 (2002).
Geibel, S., Procko, E., Hultgren, S. J., Baker, D. & Waksman, G. Structural and energetic basis of folded-protein transport by the FimD usher. Nature 496, 243–246 (2013).
Volkan, E. et al. Domain activities of PapC usher reveal the mechanism of action of an Escherichia coli molecular machine. Proc. Natl Acad. Sci. USA 109, 9563–9568 (2012).
Baga, M., Norgren, M. & Normark, S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell 49, 241–251 (1987).
Verger, D., Miller, E., Remaut, H., Waksman, G. & Hultgren, S. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 7, 1228–1232 (2006).
Nishiyama, M., Ishikawa, T., Rechsteiner, H. & Glockshuber, R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320, 376–379 (2008).
Saulino, E. T., Thanassi, D. G., Pinkner, J. S. & Hultgren, S. J. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 17, 2177–2185 (1998).
Li, Q. et al. The differential affinity of the usher for chaperone–subunit complexes is required for assembly of complete pili. Mol. Microbiol. 76, 159–172 (2010).
Jacob-Dubuisson, F., Heuser, J., Dodson, K., Normark, S. & Hultgren, S. Initiation of assembly and association of the structural elements of a bacterial pilus depend on two specialized tip proteins. EMBO J. 12, 837–847 (1993).
Lee, Y. M., Dodson, K. W. & Hultgren, S. J. Adaptor function of PapF depends on donor strand exchange in P-pilus biogenesis of Escherichia coli. J. Bacteriol. 189, 5276–5283 (2007).
Rose, R. J. et al. Unraveling the molecular basis of subunit specificity in P pilus assembly by mass spectrometry. Proc. Natl Acad. Sci. USA 105, 12873–12878 (2008).
Verger, D. et al. Structural determinants of polymerization reactivity of the P pilus adaptor subunit PapF. Structure 16, 1724–1731 (2008).
Eidam, O., Dworkowski, F. S., Glockshuber, R., Grutter, M. G. & Capitani, G. Crystal structure of the ternary FimC–FimFt–FimDN complex indicates conserved pilus chaperone–subunit complex recognition by the usher FimD. FEBS Lett. 582, 651–655 (2008).
Ng, T. W., Akman, L., Osisami, M. & Thanassi, D. G. The usher N terminus is the initial targeting site for chaperone–subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 186, 5321–5331 (2004).
Werneburg, G. T. et al. The pilus usher controls protein interactions via domain masking and is functional as an oligomer. Nat. Struct. Mol. Biol. 22, 540–546 (2015).
Pinkner, J. S. et al. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl Acad. Sci. USA 103, 17897–17902 (2006).
Henderson, N. S. & Thanassi, D. G. Purification of the outer membrane usher protein and periplasmic chaperone–subunit complexes from the P and type 1 pilus systems. Methods Mol. Biol. 966, 37–52 (2013).
Dodson, K. W. et al. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell 105, 733–743 (2001).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 (1997).
Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006).
Hayward, S. & Berendsen, H. J. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30, 144–154 (1998).
Volkan, E. et al. Molecular basis of usher pore gating in Escherichia coli pilus biogenesis. Proc. Natl Acad. Sci. USA 110, 20741–20746 (2013).
Slonim, L. N., Pinkner, J. S., Branden, C. I. & Hultgren, S. J. Interactive surface in the PapD chaperone cleft is conserved in pilus chaperone superfamily and essential in subunit recognition and assembly. EMBO J. 11, 4747–4756 (1992).
Acknowledgements
We thank the staff of beamline 24-ID-E at APS for assistance, especially K. Rajashankar, K. Perry and N. Sukumar. This work used NE-CAT beamlines (GM103403), a Pilatus detector (RR029205) and an Eiger detector (OD021527) at the APS (DE-AC02-06CH11357). This work was supported by grants from the NIH (R01AI029549 and R01AI048689 to S.J.H.) and National Science Foundation (DGE-1745038 to N.S.O.), as well as start-up funds from Washington University School of Medicine (to P.Y.).
Author information
Authors and Affiliations
Contributions
N.S.O. expressed and purified protein, and produced and crystallized the PapC PapDG complex. N.S.O. and Z.D. collected X-ray data and determined the structure. P.Y. supervised the crystallography work. J.S.P. expressed and purified protein. S.J.H., P.Y., N.S.O. and Z.D. designed the in vitro functional and biochemical assays, and N.S.O. performed them. S.J.H., P.Y., K.W.D., F.A., Z.D. and N.S.O. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1, Supplementary Figures 1–7
Rights and permissions
About this article
Cite this article
Omattage, N.S., Deng, Z., Pinkner, J.S. et al. Structural basis for usher activation and intramolecular subunit transfer in P pilus biogenesis in Escherichia coli. Nat Microbiol 3, 1362–1368 (2018). https://doi.org/10.1038/s41564-018-0255-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0255-y
This article is cited by
-
The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili
Nature Communications (2024)
-
Stochastic chain termination in bacterial pilus assembly
Nature Communications (2023)
-
Electron cryo-microscopy reveals the structure of the archaeal thread filament
Nature Communications (2022)
-
Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis
Nature Communications (2021)
-
Urinary tract infections: microbial pathogenesis, host–pathogen interactions and new treatment strategies
Nature Reviews Microbiology (2020)