Abstract
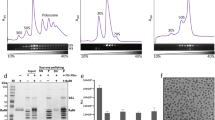
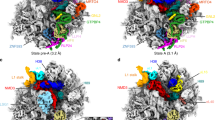
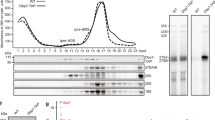
To survive under conditions of stress, such as nutrient deprivation, bacterial 70S ribosomes dimerize to form hibernating 100S particles1. In γ-proteobacteria, such as Escherichia coli, 100S formation requires the ribosome modulation factor (RMF) and the hibernation promoting factor (HPF)2,3,4. Here we present single-particle cryo-electron microscopy structures of hibernating 70S and 100S particles isolated from stationary-phase E. coli cells at 3.0 Å and 7.9 Å resolution, respectively. The structures reveal the binding sites for HPF and RMF as well as the unexpected presence of deacylated E-site transfer RNA and ribosomal protein bS1. HPF interacts with the anticodon-stem-loop of the E-tRNA and occludes the binding site for the messenger RNA as well as A- and P-site tRNAs. RMF facilitates stabilization of a compact conformation of bS1, which together sequester the anti-Shine-Dalgarno sequence of the 16S ribosomal RNA (rRNA), thereby inhibiting translation initiation. At the dimerization interface, the C-terminus of uS2 probes the mRNA entrance channel of the symmetry-related particle, thus suggesting that dimerization inactivates ribosomes by blocking the binding of mRNA within the channel. The back-to-back E. coli 100S arrangement is distinct from 100S particles observed previously in Gram-positive bacteria5,6,7,8, and reveals a unique role for bS1 in translation regulation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gohara, D. W. & Yap, M. F. Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr. Genet. 64, 753–760 (2018).
Wada, A., Yamazaki, Y., Fujita, N. & Ishihama, A. Structure and probable genetic location of a ribosome modulation factor associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc. Natl Acad. Sci. USA 87, 2657–2661 (1990).
Maki, Y., Yoshida, H. & Wada, A. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5, 965–974 (2000).
Ueta, M. et al. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells 10, 1103–1112 (2005).
Beckert, B. et al. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 36, 2061–2072 (2017).
Khusainov, I. et al. Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. EMBO J. 36, 2073–2087 (2017).
Matzov, D. et al. The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat. Commun. 8, 723 (2017).
Franken, L. E.et al. A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy. Nat. Commun. 8, 722 (2017).
Yoshida, H. & Wada, A. The 100S ribosome: ribosomal hibernation induced by stress. Wiley Interdiscip. Rev. RNA 5, 723–732 (2014).
McKay, S. L. & Portnoy, D. A. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob. Agents Chemother. 59, 6992–6999 (2015).
Harms, A., Maisonneuve, E. & Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354, 1390–1399 (2016).
Kato, T. et al. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18, 719–724 (2010).
Ortiz, J. O. et al. Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J. Cell Biol. 190, 613–621 (2010).
Vila-Sanjurjo, A., Schuwirth, B. S., Hau, C. W. & Cate, J. H. D. Structural basis for the control of translational initiation during stress. Nat. Struct. Mol. Biol. 11, 1054–1059 (2004).
Polikanov, Y. S., Blaha, G. M. & Steitz, T. A. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336, 915–918 (2012).
Dunkle, J. A. et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981–984 (2011).
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015).
Dunkle, J. A. & Cate, J. H. in Ribosomes. Structure, Function, and Dynamics (eds Wintermeyer, W., Rodnina, M.V. & Green R.) Ch. 6, 65–73 (Springer, Vienna, 2011).
Sengupta, J., Agrawal, R. & Frank, J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messengerRNA. Proc. Natl Acad. Sci. USA 98, 11991–11996 (2001).
Byrgazov, K. et al. Structural basis for the interaction of protein S1 with the Escherichia coli ribosome. Nucleic Acids Res. 43, 661–673 (2015).
Park, E. et al. Structure of the SecY channel during initiation of protein translocation. Nature 506, 102–106 (2014).
Demo, G. et al. Structure of RNA polymerase bound to ribosomal 30S subunit. eLife 6, e28560 (2017).
Salah, P. et al. Probing the relationship between Gram-negative and Gram-positive S1 proteins by sequence analysis. Nucleic Acids Res. 37, 5578–5588 (2009).
Subramanian, A.-R. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 28, 101–142 (1983).
Qu, X., Lancaster, L., Noller, H. F., Bustamante, C. & Tinoco, I.Jr.. Ribosomal protein S1 unwinds double-stranded RNA in multiple steps. Proc. Natl Acad. Sci., USA 109, 14458–14463 (2012).
Duval, M. et al. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol. 11, e1001731 (2013).
Aliprandi, P. et al. S1 ribosomal protein functions in translation initiation and ribonuclease RegB activation are mediated by similar RNA-protein interactions: an NMR and SAXS analysis. J. Biol. Chem. 283, 13289–13301 (2008).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Huter, P., Muller, C., Arenz, S., Beckert, B. & Wilson, D. N. Structural basis for ribosome rescue in bacteria. Trends Biochem. Sci. 42, 669–680 (2017).
Saguy, M., Gillet, R., Skorski, P., Hermann-Le Denmat, S. & Felden, B. Ribosomal protein S1 influences trans-translation in vitro and in vivo. Nucleic Acids Res. 35, 2368–2376 (2007).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. eLife 4, e06980 (2015).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Rosenthal, P. & Henderson, R. Optimal determination of particle orientation, absolute hand, and control loss in single particle electron microscopy. J. Mol. Biol. 333, 721–745 (2003).
Scheres, S. H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Moriya, T. et al. High-resolution single particle analysis from electron cryo-microscopy images using SPHIRE. J .Vis. Exp. 123, e55448 (2017).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D71, 136–153 (2015).
Adams, P. D. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221 (2010).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Florin, T. et al. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat. Struct. Mol. Biol. 24, 752–757 (2017).
Pettersen, E. F. et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D60, 2126–2132 (2004).
Arenz, S.et al. A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest. Nat. Commun. 7, 12026 (2016).
Giraud, P., Crechet, J. B., Uzan, M., Bontems, F. & Sizun, C. Resonance assignment of the ribosome binding domain of E. coli ribosomal protein S1. Biomol. NMR Assign. 9, 107–111 (2015).
Polikanov, Y. S., Steitz, T. A. & Innis, C. A. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 21, 787–793 (2014).
Kaminishi, T. et al. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure 15, 289–297 (2007).
Yusupova, G., Jenner, L., Rees, B., Moras, D. & Yusupov, M. Structural basis for messenger RNA movement on the ribosome. Nature 444, 391–394 (2006).
Acknowledgements
We thank S. Rieder and C. Ungewickell for expert technical assistance and C. Schmidt for IT support. This research was supported by grants from the Deutsche Forschungsgemeinschaft SPP1879 (to D.N.W), CZ234/1–1 (to A.C.), FOR1805 (to D.N.W., Z.I. and R.B.).
Author information
Authors and Affiliations
Contributions
D.N.W. designed the study. B.B. prepared the cryo-EM sample. O.B. collected the single-particle cryo-EM data, which was processed by B.B. The tilt data were collected and processed by M.T. A.C. performed the microarray analysis. B.B. built and refined the molecular models and generated the figures. B.B., M.T., R.B., Z.I., J.P. and D.N.W. interpreted the results. B.B. and D.N.W. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Data availability
Cryo-electron density maps of the hibernating 70S and 100S ribosomal particles are available through the EMDB with entry codes EMD-0137 and EMD-0139, respectively. Molecular models of the hibernating 70S and 100S ribosomal particles are deposited in the Protein Data Bank with entry code 6H4N and 6H58, respectively. The data that support the findings of this study are available from the corresponding authors on request.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–11, Supplementary Table 1.
Supplementary Video 1
Overview of the cryo-EM maps for hibernating 70S and 100S particles.
Rights and permissions
About this article
Cite this article
Beckert, B., Turk, M., Czech, A. et al. Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat Microbiol 3, 1115–1121 (2018). https://doi.org/10.1038/s41564-018-0237-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0237-0
This article is cited by
-
Structural basis of ribosomal 30S subunit degradation by RNase R
Nature (2024)
-
Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH
Nature Communications (2024)
-
Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization
Nature Communications (2023)
-
A molecular network of conserved factors keeps ribosomes dormant in the egg
Nature (2023)
-
CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state
Nature Microbiology (2023)