Abstract
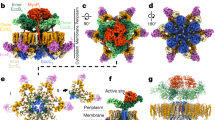
Type IV secretion systems (T4SSs) are complex machines used by bacteria to deliver protein and DNA complexes into target host cells1,2,3,4,5. Conserved ATPases are essential for T4SS function, but how they coordinate their activities to promote substrate transfer remains poorly understood. Here, we show that the DotB ATPase associates with the Dot–Icm T4SS at the Legionella cell pole through interactions with the DotO ATPase. The structure of the Dot–Icm apparatus was solved in situ by cryo-electron tomography at 3.5 nm resolution and the cytoplasmic complex was solved at 3.0 nm resolution. These structures revealed a cell envelope-spanning channel that connects to the cytoplasmic complex. Further analysis revealed a hexameric assembly of DotO dimers associated with the inner membrane complex, and a DotB hexamer associated with the base of this cytoplasmic complex. The assembly of a DotB–DotO energy complex creates a cytoplasmic channel that directs the translocation of substrates through the T4SS. These data define distinct stages in Dot–Icm machine biogenesis, advance our understanding of channel activation, and identify an envelope-spanning T4SS channel.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Christie, P. J., Whitaker, N. & Gonzalez-Rivera, C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 1843, 1578–1591 (2014).
Gonzalez-Rivera, C., Bhatty, M. & Christie, P. J. Mechanism and function of type IV secretion during infection of the human host. Microbiol. Spectr. 4, 1–29 (2016).
Costa, T. R. et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359 (2015).
Trokter, M., Felisberto-Rodrigues, C., Christie, P. J. & Waksman, G. Recent advances in the structural and molecular biology of type IV secretion systems. Curr. Opin. Struct. Biol. 27, 16–23 (2014).
Kubori, T. & Nagai, H. The type IVB secretion system: an enigmatic chimera. Curr. Opin. Microbiol. 29, 22–29 (2015).
Low, H. H. et al. Structure of a type IV secretion system. Nature 508, 550–553 (2014).
Kubori, T. et al. Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proc. Natl Acad. Sci. USA 111, 11804–11809 (2014).
Ghosal, D., Chang, Y. W., Jeong, K. C., Vogel, J. P. & Jensen, G. J. In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep. 18, 726–732 (2017).
Jeong, K. C., Ghosal, D., Chang, Y. W., Jensen, G. J. & Vogel, J. P. Polar delivery of Legionella type IV secretion system substrates is essential for virulence. Proc. Natl Acad. Sci. USA 114, 8077–8082 (2017).
Vincent, C. D. et al. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 62, 1278–1291 (2006).
Sexton, J. A., Yeo, H. J . & Vogel, J. P. Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol. Microbiol. 57, 70–84 (2005).
Sutherland, M. C., Nguyen, T. L., Tseng, V. & Vogel, J. P. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Path 8, e1002910 (2012).
Vincent, C. D., Friedman, J. R., Jeong, K. C., Sutherland, M. C. & Vogel, J. P. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol 85, 378–391 (2012).
Kwak, M. J. et al. Architecture of the type IV coupling protein complex of Legionella pneumophila . Nat. Microbiol. 2, 17114 (2017).
Dang, T. A. & Christie, P. J. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J. Bacteriol. 179, 453–462 (1997).
Dang, T. A., Zhou, X. R., Graf, B. & Christie, P. J. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol. Microbiol 32, 1239–1253 (1999).
Durand, E., Oomen, C. & Waksman, G. Biochemical dissection of the ATPase TraB, the VirB4 homologue of the Escherichia coli pKM101 conjugation machinery. J. Bacteriol. 192, 2315–2323 (2010).
Durand, E., Waksman, G. & Receveur-Brechot, V. Structural insights into the membrane-extracted dimeric form of the ATPase TraB from the Escherichia coli pKM101 conjugation system. BMC Struct. Biol. 11, 4 (2011).
Wallden, K. et al. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc. Natl Acad. Sci. USA 109, 11348–11353 (2012).
Roy, C. R. & Isberg, R. R. Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infect. Immun. 65, 571–578 (1997).
Buscher, B. A. et al. The DotL protein, a member of the TraG-coupling protein family, is essential for viability of Legionella pneumophila strain Lp02. J. Bacteriol. 187, 2927–2938 (2005).
Merriam, J. J., Mathur, R., Maxfield-Boumil, R. & Isberg, R. R. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65, 2497–2501 (1997).
Chetrit, D. et al. Negative regulation of the endocytic adaptor disabled-2 (Dab2) in mitosis. J. Biol. Chem. 286, 5392–5403 (2011).
Zuckman, D. M., Hung, J. B. & Roy, C. R. Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol. Microbiol. 32, 990–1001 (1999).
Hu, B., Lara-Tejero, M., Kong, Q., Galan, J. E. & Liu, J. In situ molecular architecture of the Salmonella type III secretion machine. Cell 168, 1065–1074 (2017).
Morado, D. R., Hu, B. & Liu, J. Using Tomoauto: A protocol for high-throughput automated cryo-electron tomography. J. Vis. Exp. 107, e53608 (2016).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Agulleiro, J. I. & Fernandez, J. J. Tomo3D 2.0—exploitation of advanced vector extensions (AVX) for 3D reconstruction. J. Struct. Biol. 189, 147–152 (2015).
Hu, B. et al. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc. Natl Acad. Sci. USA 112, 1047–1052 (2015).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Agard, D. A., Hiraoka, Y., Shaw, P. & Sedat, J. W. Fluorescence microscopy in three dimensions. Methods Cell Biol. 30, 353–377 (1989).
Frick-Cheng, A. E. et al. Molecular and structural analysis of the Helicobacter pylori cag type IV secretion system core complex. mBio 7, e02001–e02015 (2016).
Berger, K. H. & Isberg, R. R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol 7, 7–19 (1993).
Andrews, H. L., Vogel, J. P. & Isberg, R. R. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66, 950–958 (1998).
Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876 (1998).
Finan, T. M., Kunkel, B., De Vos, G. F. & Signer, E. R. Second symbiotic megaplasmid in R hizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167, 66–72 (1986).
Acknowledgements
B.H. and J.L were supported by the National Institutes of Health (R01AI087946 and R01GM107629) and the Welch Foundation (AU-1714). D.C. and C.R. were supported by the NIH (R37AI041699 and R21AI130671). P.C. was supported by the NIH (R01GM48476). We are grateful to S.S. Ivanov (Louisiana State University) for insightful suggestions and critique; H. Nagai (Gifu University) for the L. pneumophila ΔT4SS strain; R.R. Isberg (Tufts University) for the antibody to DotO; E.H. Rego (Yale University) for technical assistance with FRAP.
Author information
Authors and Affiliations
Contributions
B.H., C.R., D.C. and J.L. designed research. D.C. constructed the L. pneumophila expression plasmids and strains. B.H. and D.C. collected and together with C.R., J.L. and P.C. analysed the data. B.H., C.R., D.C., J.L. and P.C. wrote the paper.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–8, and Supplementary Tables 1 and 2.
Supplementary Video 1
Real-time visualization of DotB–sfGFP, sfGFP, and DotBE191K–sfGFP expressed from dot dotB operon in Legionella pneumophila.
Supplementary Video 2
3D visualization of a tomographic reconstruction and the intact T4SS machine
Rights and permissions
About this article
Cite this article
Chetrit, D., Hu, B., Christie, P.J. et al. A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat Microbiol 3, 678–686 (2018). https://doi.org/10.1038/s41564-018-0165-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0165-z
This article is cited by
-
Uncovering microbiomes of the rice phyllosphere using long-read metagenomic sequencing
Communications Biology (2024)
-
Structural and functional diversity of type IV secretion systems
Nature Reviews Microbiology (2024)
-
Cytosolic sorting platform complexes shuttle type III secretion system effectors to the injectisome in Yersinia enterocolitica
Nature Microbiology (2024)
-
Cryo-EM structure of a type IV secretion system
Nature (2022)
-
Pseudomonas putida mediates bacterial killing, biofilm invasion and biocontrol with a type IVB secretion system
Nature Microbiology (2022)