Abstract
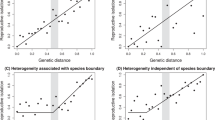
Speciation can involve a transition from a few genetic loci that are resistant to gene flow to genome-wide differentiation. However, only limited data exist concerning this transition and the factors promoting it. Here, we study phases of speciation using data from >100 populations of 11 species of Timema stick insects. Consistent with early phases of genic speciation, adaptive colour-pattern loci reside in localized genetic regions of accentuated differentiation between populations experiencing gene flow. Transitions to genome-wide differentiation are also observed with gene flow, in association with differentiation in polygenic chemical traits affecting mate choice. Thus, intermediate phases of speciation are associated with genome-wide differentiation and mate choice, but not growth of a few genomic islands. We also find a gap in genomic differentiation between sympatric taxa that still exchange genes and those that do not, highlighting the association between differentiation and complete reproductive isolation. Our results suggest that substantial progress towards speciation may involve the alignment of multi-faceted aspects of differentiation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Seehausen, O. et al. Genomics and the origin of species. Nat. Rev. Genet. 15, 176–192 (2014).
Coyne, J. A. & Orr, H. A. Speciation 1st edn (Sinauer Associates, 2004).
Flaxman, S., Walchoder, A., Feder, J. L. & Nosil, P. Theoretical models of the influence of genomic architecture on speciation. Mol. Ecol. 23, 4074–4088 (2014).
Wu, C. The genic view of the process of speciation. J. Evol. Biol. 14, 851–865 (2001).
Mallet, J. A species definition for the modern synthesis. Trends Ecol. Evol. 10, 294–299 (1995).
Feder, J. L., Egan, S. P. & Nosil, P. The genomics of speciation-with-gene-flow. Trends Genet. 28, 342–350 (2012).
Poelstra, J. W. et al. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science 344, 1410–1414 (2014).
Nadeau, N. J. et al. Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato . Genome Res. 24, 1316–1333 (2014).
Nadeau, N. J. et al. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Phil. Trans. R. Soc. B 367, 343–353 (2012).
Michel, A. P. et al. Widespread genomic divergence during sympatric speciation. Proc. Natl Acad. Sci. USA 107, 9724–9729 (2010).
Nosil, P., Egan, S. P. & Funk, D. J. Heterogeneous genomic differentiation between walking-stick ecotypes: “isolation by adaptation” and multiple roles for divergent selection. Evolution 62, 316–336 (2008).
Shafer, A. B. A. & Wolf, J. B. W. Widespread evidence for incipient ecological speciation: a meta-analysis of isolation-by-ecology. Ecol. Lett. 16, 940–950 (2013).
Soria-Carrasco, V. et al. Stick insect genomes reveal natural selection’s role in parallel speciation. Science 344, 738–742 (2014).
Egan, S. P. et al. Experimental evidence of genome-wide impact of ecological selection during early stages of speciation-with-gene-flow. Ecol. Lett. 18, 817–825 (2015).
Burke, M. K. How does adaptation sweep through the genome? Insights from long-term selection experiments. Proc. R. Soc. B 279, 5029–5038 (2012).
Lamichhaney, S. et al. Population-scale sequencing reveals genetic differentiation due to local adaptation in Atlantic herring. Proc. Natl Acad. Sci. USA 109, 19345–19350 (2012).
Lawniczak, M. K. N. et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330, 512–514 (2010).
Gavrilets, S. Fitness Landscapes and the Origin of Species Vol. 41 (Princeton Univ. Press, 2004).
Barton, N. H. Multilocus clines. Evolution 37, 454–471 (1983).
Kirkpatrick, M. & Ravigné, V. Speciation by natural and sexual selection: models and experiments. Am. Nat. 159, S22–S35 (2002).
Jiggins, C. D. & Mallet, J. Bimodal hybrid zones and speciation. Trends Ecol. Evol. 15, 250–255 (2000).
Nosil, P. Ecological Speciation (Oxford Univ. Press, 2012).
Turner, T. L. & Hahn, M. W. Genomic islands of speciation or genomic islands and speciation? Mol. Ecol. 19, 848–850 (2010).
Turner, T. L., Hahn, M. W. & Nuzhdin, S. V. Genomic islands of speciation in Anopheles gambiae . PLoS Biol. 3, 1572–1578 (2005).
Mayr, E. Animal Species and Evolution (Harvard Univ. Press, 1963).
Yeaman, S., Aeschbacher, S. & Burger, R. The evolution of genomic islands by increased establishment probability of linked alleles. Mol. Ecol. 25, 2542–2558 (2016).
Brawand, D. et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513, 375–381 (2014).
Feulner, P. G. D. et al. Genomics of divergence along a continuum of parapatric population differentiation. PLoS Genet. 11, e1004966 (2015).
Burri, R. et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 25, 1656–1665 (2015).
Martin, S. H. et al. Genome-wide evidence for speciation with gene flow in Heliconius butterflies Genome Res. 23, 1817–1828 (2013).
Cruickshank, T. E. & Hahn, M. W. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol. 23, 3133–3157 (2014).
Darwin, C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life (John Murray, 1859).
Peccoud, J., Ollivier, A., Plantegenest, M. & Simon, J. C. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl Acad. Sci. USA 106, 7495–7500 (2009).
Mallet, J., Beltran, M., Neukirchen, W. & Linares, M. Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol. Biol. 7, 28 (2007).
Orr, H. A. The population-genetics of speciation—the evolution of hybrid incompatibilities. Genetics 139, 1805–1813 (1995).
Law, J. H. & Crespi, B. J. The evolution of geographic parthenogenesis in Timema walking-sticks. Mol. Ecol. 11, 1471–1489 (2002).
Nosil, P. Divergent host plant adaptation and reproductive isolation between ecotypes of Timema cristinae walking sticks. Am. Nat. 169, 151–162 (2007).
Nosil, P. & Sandoval, C. P. Ecological niche dimensionality and the evolutionary diversification of stick insects. PLoS ONE 3, e1907 (2008).
Nosil, P. et al. Genomic consequences of multiple speciation processes in a stick insect. Proc. R. Soc. B 279, 5058–5065 (2012).
Hofer, T., Foll, M. & Excoffier, L. Evolutionary forces shaping genomic islands of population differentiation in humans. BMC Genomics 13, 107 (2012).
Sandoval, C. P. The effects of relative geographical scales of gene flow and selection on morph frequencies in the walking-stick Timema cristinae . Evolution 48, 1866–1879 (1994).
Comeault, A. A. et al. Selection on a genetic polymorphism counteracts ecological speciation in a stick insect. Curr. Biol. 25, 1–7 (2015).
Nosil, P. & Hohenlohe, P. A. Dimensionality of sexual isolation during reinforcement and ecological speciation in Timema cristinae stick insects. Evol. Ecol. Res. 14, 467–485 (2012).
Nosil, P. & Crespi, B. J. Does gene flow constrain adaptive divergence or vice versa? A test using ecomorphology and sexual isolation in Timema cristinae walking-sticks. Evolution 58, 102–112 (2004).
Schwander, T. et al. Hydrocarbon divergence and reproductive isolation in Timema stick insects. BMC Evol. Biol. 13, 151 (2013).
Chung, H. et al. A single gene affects both ecological divergence and mate choice in Drosophila . Science 343, 1148–1151 (2014).
Yang, J. et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43, 519–525 (2011).
Zhou, X., Carbonetto, P. & Stephens, M. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 9, e1003264 (2013).
Arbuthnott, D. & Crespi, B. J. Courtship and mate discrimination within and between species of Timema walking-sticks. Anim. Behav. 78, 53–59 (2009).
Grant, B. R. & Grant, P. R. Fission and fusion of Darwin’s finches populations. Phil. Trans. R. Soc. B 363, 2821–2829 (2008).
Riesch, R., Barrett-Lennard, L. G., Ellis, G. M., Ford, J. K. B. & Deecke, V. B. Cultural traditions and the evolution of reproductive isolation: ecological speciation in killer whales? Biol. J. Linn. Soc. 106, 1–17 (2012).
Wood, T. K. & Keese, M. C. Host-plant induced assortative mating in Enchenopa treehoppers. Evolution 44, 619–628 (1990).
Gompert, Z. et al. Admixture and the organization of genetic diversity in a butterfly species complex revealed through common and rare genetic variants. Mol. Ecol. 23, 4555–4573 (2014).
Cummings, M. P., Neel, M. C. & Shaw, K. L. A genealogical approach to quantifying lineage divergence. Evolution 62, 2411–2422 (2008).
Buerkle, C. A. & Gompert, Z. Population genomics based on low coverage sequencing: How low should we go? Mol. Ecol. 22, 3028–3035 (2013).
Fumagalli, M. et al. Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195, 979–992 (2013).
Barton, N. H. & Gale, K. S. in Hybrid Zones and the Evolutionary Process (ed. Harrison, R. G. ) 13–45 (Oxford Univ. Press, 1993).
Derryberry, E. P., Derryberry, G. E., Maley, J. M. & Brumfield, R. T. hzar: hybrid zone analysis using an R software package. Mol. Ecol. Resour. 14, 652–663 (2014).
Weir, B. S. & Cockerham, C. C. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984).
Baum, L. E., Petrie, T., Soules, G. & Weiss, N. A maximization technique occurring in statistical analysis of probabilistic functions of Markov chains. Ann. Math. Stat. 41, 164–171 (1970).
Harte, D. HiddenMarkov: Hidden Markov Models. R package version 1.7-0 (Statistics Research Associates, accessed 14 April 2012); http://cran.at.r-project.org/web/packages/HiddenMarkov
Harte, D. HiddenMarkov: Hidden Markov Models. R package version 1.8-3 (Statistics Research Associates, accessed 13 April 2015); http://cran.at.r-project.org/web/packages/HiddenMarkov
Gompert, Z. et al. Experimental evidence for ecological selection on genome variation in the wild. Ecol. Lett. 17, 369–379 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Blows, M. W. & Allan, R. A. Levels of mate recognition within and between two Drosophila species and their hybrids. Am. Nat. 152, 826–837 (1998).
Rundle, H. D., Chenoweth, S. F., Doughty, P. & Blows, M. W. Divergent selection and the evolution of signal traits and mating preferences. PLoS Biol. 3, e368 (2005).
Aitchison, J. The Statistical Analysis of Compositional Data 12th edn (Chapman & Hall, 1986).
Wray, N. R., Yang, J., Goddard, M. E. & Visscher, P. M. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 6, e1000864 (2010).
Nosil, P., Crespi, B. J. & Sandoval, C. P. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature 417, 440–443 (2002).
Rolan-Alvarez, E. & Caballero, M. Estimating sexual selection and sexual isolation effects from mating frequencies. Evolution 54, 30–36 (2000).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H., The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Hudson, R. R., Slatkin, M. & Maddison, W. P. Estimation of levels of gene flow from DNA-sequence data. Genetics 132, 583–589 (1992).
Bhatia, G., Patterson, N., Sankararaman, S. & Price, A. L. Estimating and interpreting Fst: the impact of rare variants. Genome Res. 23, 1514–1521 (2013).
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011).
Clarke, R. T., Rothery, P. & Raybould, A. F. Confidence limits for regression relationships between distance matrices: estimating gene flow with distance. J. Agric. Biol. Env. Stat. 7, 361–372 (2002).
Nei, M. Molecular Evolutionary Genetics (Columbia Univ. Press, 1987).
Wright, S. Isolation by distance. Genetics 28, 114–138 (1943).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Skotte, L., Korneliussen, T. S. & Albrechtsen, A. Estimating individual admixture proportions from next generation sequencing data. Genetics 195, 693–702 (2013).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Stamatakis, A., Hoover, P. & Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771 (2008).
Abràmoff, M. D., Magalhães, P. J. & Ram, S. J. Image processing with ImageJ. Biophoton. Int. 11, 36–42 (2004).
Endler, J. A. A framework for analysing colour pattern geometry: adjacent colours. Biol. J. Linn. Soc. 107, 233–253 (2012).
Beuttell, K. & Losos, J. B. Ecological morphology of Caribbean anoles. Herpetol. Monogr. 13, 1–28 (1999).
Bouckaert, R., Alvarado-Mora, M. V. & Pinho, J. R. R. Evolutionary rates and HBV: issues of rate estimation with Bayesian molecular methods. Antivir. Ther. 18, 497–503 (2013).
Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014).
Acknowledgements
We thank R. Guillem, R. Kather, J. Stapley and T. Schwander for their advice; the Oakley-, Kuris- and Lafferty-groups at University of California Santa Barbara for their support; L. Jeanson and J. Hosegood for the phenotypic measurements and lab work; R. Marin for drawing all the figures; T. Schwander for providing the genetic crosses used for linkage mapping; the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z and MRC Hub grant G0900747 91070) for generating the whole-genome re-sequencing data; and the National Center for Genome Sequencing (USA) for the GBS data. R.R. was supported by the Human Frontier Science Program, M.M. and K.L. were supported by the Swiss National Science Foundation and P.N. was supported by the Royal Society of London. The work was funded by grants from the European Research Council (grant NatHisGen R/129639 to P.N.) and the Natural Science and Engineering Research Council of Canada (to B.J.C. and G.G.). Computing, storage and other resources from the Division of Research Computing in the Office of Research and Graduate Studies at Utah State University, as well as access to the High Performance Computing Facilities, particularly to the Iceberg HPC cluster, from the Corporate Information and Computing Services at the University of Sheffield, are gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
R.R., Z.G., M.M., G.G., J.F., B.J.C. and P.N. conceived the project. R.R., R.V., D.L., M.M., A.A.C., R.G., T.E.F., C.P.S., C.F.d.C. and P.N. collected the data. R.R., D.L., M.M., R.V., Z.G. and P.N. led the data analyses, aided by V.S.-C., K.L., C.F.d.C. and S.R.D. R.R., Z.G. and P.N. wrote the initial manuscript and all authors contributed to further writing and revisions. Z.G. and V.S.-C. organized the data archiving. R.R., M.M., D.L., R.V. and Z.G. contributed equally.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests
Supplementary information
Supplementary Information
Supplementary Methods; Supplementary Tables 1–15; Supplementary Figures 1–4 (PDF 3977 kb)
Rights and permissions
About this article
Cite this article
Riesch, R., Muschick, M., Lindtke, D. et al. Transitions between phases of genomic differentiation during stick-insect speciation. Nat Ecol Evol 1, 0082 (2017). https://doi.org/10.1038/s41559-017-0082
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-017-0082
This article is cited by
-
High disparity in repellent gland anatomy across major lineages of stick and leaf insects (Insecta: Phasmatodea)
BMC Zoology (2024)
-
Divergent dynamics of sexual and habitat isolation at the transition between stick insect populations and species
Nature Communications (2024)
-
Life on a beach leads to phenotypic divergence despite gene flow for an island lizard
Communications Biology (2023)
-
Early stages of sympatric homoploid hybrid speciation in crater lake cichlid fishes
Nature Communications (2022)
-
Climatic similarity and genomic background shape the extent of parallel adaptation in Timema stick insects
Nature Ecology & Evolution (2022)