Abstract
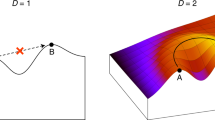
The adaptive landscape is an iconic metaphor that pervades evolutionary biology. It was mostly applied in theoretical models until recent years, when empirical data began to allow partial landscape reconstructions. Here, we exhaustively analyse 1,137 complete landscapes from 129 eukaryotic species, each describing the binding affinity of a transcription factor to all possible short DNA sequences. We find that the navigability of these landscapes through single mutations is intermediate to that of additive and shuffled null models, suggesting that binding affinity—and thereby gene expression—is readily fine-tuned via mutations in transcription factor binding sites. The landscapes have few peaks that vary in their accessibility and in the number of sequences they contain. Binding sites in the mouse genome are enriched in sequences found in the peaks of especially navigable landscapes and the genetic diversity of binding sites in yeast increases with the number of sequences in a peak. Our findings suggest that landscape navigability may have contributed to the enormous success of transcriptional regulation as a source of evolutionary adaptations and innovations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wright, S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. in Proc. Sixth Int. Congress Genetics Vol. 1 (ed. Jones, D. F. ) 356–366 (The Genetics Society of America, 1932).
Szendro, I. G., Schenk, M. F., Franke, J., Krug, J. & de Visser, J. A. G. M. Quantitative analyses of empirical fitness landscapes. J. Stat. Mech-Theory E. 2013, P01005 (2013).
Kauffman, S. & Levin, S. Towards a general theory of adaptive walks on rugged landscapes. J. Theor. Biol. 128, 11–45 (1987).
Rowe, W. et al. Analysis of a complete DNA-protein affinity landscape. J. R. Soc. Interface 7, 397–408 (2010).
Jiménez, J. I., Xulvi-Brunet, R., Campbell, G. W., Turk-MacLeod, R. & Chen, I. A. Comprehensive experimental fitness landscape and evolutionary network for small RNA. Proc. Natl Acad. Sci. USA 110, 14984–14989 (2013).
Wray, G. A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216 (2007).
Gertz, J., Siggia, E. D. & Cohen, B. A. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457, 215–218 (2009).
Shultzaberger, R. K., Malashock, D. S., Kirsch, J. F. & Eisen, M. B. The fitness landscapes of cis-acting binding sites in different promoter and environmental contexts. PLoS Genet. 6, e1001042 (2010).
Sharon, E. et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 30, 521–530 (2012).
Gerland, U. & Hwa, T. On the selection and evolution of regulatory DNA motifs. J. Mol. Evol. 55, 386–400 (2002).
Berg, J., Willmann, S. & Lässig, M. Adaptive evolution of transcription factor binding sites. BMC Evol. Biol. 4, 42 (2004).
Maerkl, S. J. & Quake, S. R. A systems approach to measuring the binding energy landscapes of transcription factors. Science 315, 233–237 (2007).
Mustonen, V., Kinney, J., Callan, C. G. & Lässig, M. Energy-dependent fitness: a quantitative model for the evolution of yeast transcription factor binding sites. Proc. Natl Acad. Sci. USA 105, 12376–12381 (2008).
Haldane, A., Manhart, M. & Morozov, A. V. Biophysical fitness landscapes for transcription factor binding sites. PLoS Comput. Biol. 10, e1003683 (2014).
Carlson, C. D. et al. Specificity landscapes of DNA binding molecules elucidate biological function. Proc. Natl Acad. Sci. USA 107, 4544–4549 (2010).
Weghorn, D. & Lässig, M. Fitness landscape for nucleosome positioning. Proc. Natl Acad. Sci. USA 110, 10988–10993 (2013).
Buenrostro, J. D. et al. Quantitative analysis of RNA–protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat. Biotechnol. 32, 562–568 (2014).
Newburger, D. E. & Bulyk, M. L. UniPROBE: an online database of protein binding microarray data on protein–DNA interactions. Nucleic Acids Res. 37, D77–D82 (2009).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
Badis, G. et al. Diversity and complexity in DNA recognition by transcription factors. Science 324, 1720–1723 (2009).
Berger, M. F. et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 24, 1429–1435 (2006).
Payne, J. L. & Wagner, A. The robustness and evolvability of transcription factor binding sites. Science 343, 875–877 (2014).
Zhu, C. et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 19, 556–566 (2009).
Nakagawa, S., Gisselbrecht, S. S., Rogers, J. M., Hartl, D. L. & Bulyk, M. L. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl Acad. Sci. USA 110, 12349–12354 (2013).
Maynard Smith, J. Natural selection and the concept of a protein space. Nature 225, 563–564 (1970).
Lehner, B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 27, 323–331 (2011).
Poelwijk, F. J., Tănase-Nicola, S., Kiviet, D. J. & Tans, S. J. Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. J. Theor. Biol. 272, 141–144 (2011).
Jolma, A. et al. DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013).
Weinreich, D. M., Delaney, N. F., Depristo, M. A. & Hartl, D. L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006).
Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 (2014).
Stergachis, A. B. et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515, 365–370 (2014).
Hesselberth, J. R. et al. Global mapping of protein–DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289 (2009).
Lynch, M. & Hagner, K. Evolutionary meandering of intermolecular interactions along the drift barrier. Proc. Natl Acad. Sci. USA 112, E30–E38 (2015).
MacArthur, S. & Brookfield, J. F. Y. Expected rates and modes of evolution of enhancer sequences. Mol. Biol. Evol. 21, 1064–1073 (2004).
Bergström, A. et al. A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 31, 872–888 (2014).
MacIsaac, K. D. et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae . BMC Bioinformatics 7, 113 (2006).
Gompel, N., Prud’homme, B., Wittkopp, P. J., Kassner, V. A. & Carroll, S. B. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila . Nature 433, 481–487 (2005).
Rister, J. et al. Single-base pair differences in a shared motif determine differential Rhodopsin expression. Science 350, 1258–1261 (2015).
Siggers, T. & Gordân, R. Protein–DNA binding: complexities and multi-protein codes. Nucleic Acids Res. 42, 2099–2111 (2014).
Li, X. Y. et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6, 0365–0388 (2008).
Fisher, W. W. et al. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila . Proc. Natl Acad. Sci. USA 109, 21330–21335 (2012).
Mustonen, V. & Lässig, M. From fitness landscapes to seascapes: non-equilibrium dynamics of selection and adaptation. Trends Genet. 25, 111–119 (2009).
Arbiza, L. et al. Genome-wide inference of natural selection on human transcription factor binding sites. Nat. Genet. 45, 723–729 (2013).
Mustonen, V. & Lässig, M. Evolutionary population genetics of promoters: predicting binding sites and functional phylogenies. Proc. Natl Acad. Sci. USA 102, 15936–15941 (2005).
Swanson, C. I., Schwimmer, D. B. & Barolo, S. Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr. Biol. 21, 1186–1196 (2011).
Grönlund, A., Lötstedt, P. & Elf, J. Transcription factor binding kinetics constrain noise suppression via negative feedback. Nat. Commun. 4, 1864 (2013).
Ramos, A. I. & Barolo, S. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Phil. Trans. R. Soc. B. 368, 20130018 (2013).
Crocker, J. et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160, 191–203 (2015).
Berger, M. F. & Bulyk, M. L. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 4, 393–411 (2009).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
van Helden, J., André, B. & Collado-Vides, J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 281, 827–842 (1998).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Dawid, A., Kiviet, D. J., Kogenaru, M., de Vos, M. & Tans, S. J. Multiple peaks and reciprocal sign epistasis in an empirically determined genotype-phenotype landscape. Chaos 20, 26105 (2010).
Poelwijk, F. J., Kiviet, D. J., Weinreich, D. M. & Tans, S. J. Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445, 383–386 (2007).
Franke, J., Klözer, A., de Visser, J. A. G. M. & Krug, J. Evolutionary accessibility of mutational pathways. PLoS Comput. Biol. 7, e1002134 (2011).
Parker, D. S., White, M. A., Ramos, A. I., Cohen, B. A. & Barolo, S. The cis-regulatory logic of Hedgehog gradient responses: key roles for gli binding affinity, competition, and cooperativity. Sci. Signal. 4, ra38 (2011).
Zhao, Y. & Stormo, G. D. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat. Biotechnol. 29, 480–483 (2011).
Morris, Q., Bulyk, M. L. & Hughes, T. R. Jury remains out on simple models of transcription factor specificity. Nat. Biotechnol. 29, 483–484 (2011).
Weinreich, D. M., Watson, R. A. & Chao, L. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59, 1165–1174 (2005).
Acknowledgements
J.A.-R. and J.L.P. acknowledge support through the Forschungskredit program of the University of Zurich (grant numbers FK-14-076 and K-74301-04-01). J.L.P. acknowledges additional support through the Ambizione program of the Swiss National Science Foundation. A.W. acknowledges support through the Swiss National Science Foundation (grant 31003A_146137) and the University Priority Research Program in Evolutionary Biology at the University of Zurich. We thank S. Bratulic, F. Khalid, A. Moya, Y. Schaerli and M. Toll-Riera for discussions and helpful comments on this manuscript.
Author information
Authors and Affiliations
Contributions
J.A.-R., J.L.P. and A.W. designed the research. J.A.-R. and J.L.P. performed the research. J.A.-R., J.L.P. and A.W. analysed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–33; Supplementary Analyses, Results and Discussion (PDF 1981 kb)
Supplementary Table 1
Information about the 1,137 transcription factors and their landscapes. (XLSX 212 kb)
Supplementary Table 2
Sources for the DNase I hypersensitive regions, footprints, and RNA-seq data for the 14 cell and tissue types analysed in this study.Sources for the DNase I hypersensitive regions, footprints, and RNA-seq data for the 14 cell and tissue types analysed in this study. (XLSX 10 kb)
Supplementary Table 3
Spearman’s rank correlations between peak accessibility and the abundance of the highest-affinity site in DNase I footprints for the 14 cell and tissue types, in addition to Wilcoxon rank-sum tests comparing binding site abundance amongst single-peaked and multi-peaked landscapes. (XLSX 54 kb)
Supplementary Table 4
Spearman’s rank correlations between peak accessibility and the abundance of the highest-affinity site in DNase hypersensitive regions for 14 murine cell and tissue types, after having removed the DNase I footprints, in addition to Wilcoxon rank-sum tests comparing binding site abundance in the same hypersensitive regions amongst single-peaked and multi-peaked landscapes. (XLSX 57 kb)
Supplementary Table 5
Analysis of covariance (ANCOVA) and its underlying assumptions. (XLSX 53 kb)
Supplementary Table 6
Partial Spearman’s rank correlations with binding affinity between peak accessibility and the abundance of the highest-affinity site in DNase I footprints for the 14 cell and tissue types. (XLSX 41 kb)
Supplementary Table 7
Partial Spearman’s rank correlations with PWM information content between peak accessibility and the abundance of the highest-affinity site in DNase I footprints for 14 murine cell and tissue types. (XLSX 51 kb)
Rights and permissions
About this article
Cite this article
Aguilar-Rodríguez, J., Payne, J. & Wagner, A. A thousand empirical adaptive landscapes and their navigability. Nat Ecol Evol 1, 0045 (2017). https://doi.org/10.1038/s41559-016-0045
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-016-0045
This article is cited by
-
Evolvability-enhancing mutations in the fitness landscapes of an RNA and a protein
Nature Communications (2023)
-
The structure of genotype-phenotype maps makes fitness landscapes navigable
Nature Ecology & Evolution (2022)
-
Relation Between the Number of Peaks and the Number of Reciprocal Sign Epistatic Interactions
Bulletin of Mathematical Biology (2022)
-
Towards an engineering theory of evolution
Nature Communications (2021)
-
Minimum epistasis interpolation for sequence-function relationships
Nature Communications (2020)