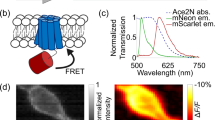
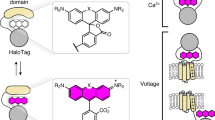
Abstract
Membrane potential is a key aspect of cellular signalling and is dynamically regulated by an array of ion-selective pumps and channels. Fluorescent voltage indicators enable non-invasive optical recording of the cellular membrane potential with high spatial resolution. Here, we report a palette of bright and sensitive hybrid voltage indicators (HVIs) with fluorescence intensities sensitive to changes in membrane potential via electrochromic Förster resonance energy transfer. Enzyme-mediated site-specific incorporation of a probe, followed by an inverse-electron-demand Diels–Alder cycloaddition, was used to create enhanced voltage-sensing rhodopsins with hybrid dye–protein architectures. The most sensitive indicator, HVI-Cy3, displays high voltage sensitivity (−39% ΔF/F0 per 100 mV) and millisecond response kinetics, enabling optical recording of action potentials at a sampling rate of 400 Hz over 10 min across a large neuronal population. The far-red indicator HVI-Cy5 could be paired with optogenetic actuators and green/red-emitting fluorescent indicators, allowing an all-optical investigation of neuronal electrophysiology.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All relevant data presented in this study are provided in the Article, Extended Data figures and Supplementary Information. The data and genetic constructs are also available from the corresponding authors upon request.
Code availability
MATLAB code can be downloaded from GitHub at https://github.com/PKUCHEMZouLab/HVI. Alternatively, it is available from the corresponding authors upon request.
References
Xu, Y. X., Zou, P. & Cohen, A. E. Voltage imaging with genetically encoded indicators. Curr. Opin. Chem. Biol. 39, 1–10 (2017).
Gong, Y. Y. et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350, 1361–1366 (2015).
Kannan, M. et al. Fast, in vivo voltage imaging using a red fluorescent indicator. Nat. Methods 15, 1108–1116 (2018).
Jin, L. et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron 75, 779–785 (2012).
Platisa, J., Vasan, G., Yang, A. & Pieribone, V. A. Directed evolution of key residues in fluorescent protein inverses the polarity of voltage sensitivity in the genetically encoded indicator ArcLight. ACS Chem. Neurosci. 8, 513–523 (2017).
St-Pierre, F. et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 17, 884–889 (2014).
Abdelfattah, A. S. et al. A bright and fast red fluorescent protein voltage indicator that reports neuronal activity in organotypic brain slices. J. Neurosci. 38, 3147–3148 (2018).
Yang, H. H. et al. Subcellular imaging of voltage and calcium signals reveals neural processing in vivo. Cell 166, 245–257 (2016).
Hou, J. H., Kralj, J. M., Douglass, A. D., Engert, F. & Cohen, A. E. Simultaneous mapping of membrane voltage and calcium in zebrafish heart in vivo reveals chamber-specific developmental transitions in ionic currents. Front. Physiol. 5, 344 (2014).
Hochbaum, D. R. et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 11, 825–833 (2014).
Fan, L. Z. et al. All-optical synaptic electrophysiology probes mechanism of ketamine-induced disinhibition. Nat. Methods 15, 823–831 (2018).
Adam, Y. et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413–417 (2019).
Flytzanis, N. C. et al. Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditis elegans neurons. Nat. Commun. 5, 4894 (2014).
Piatkevich, K. D. et al. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 14, 352–360 (2018).
Zou, P. et al. Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat. Commun. 5, 4625 (2014).
Owen, S. F., Liu, M. H. & Kreitzer, A. C. Thermal constraints on in vivo optogenetic manipulations. Nat. Neurosci. 22, 1061–1065 (2019).
Huang, Y. L., Walker, A. S. & Miller, E. W. A photostable silicon rhodamine platform for optical voltage sensing. J. Am. Chem. Soc. 137, 10767–10776 (2015).
Fluhler, E., Burnham, V. G. & Loew, L. M. Spectra, membrane binding, and potentiometric responses of new charge shift probes. Biochemistry 24, 5749–5755 (1985).
Treger, J. S., Priest, M. F., Iezzi, R. & Bezanilla, F. Real-time imaging of electrical signals with an infrared FDA-approved dye. Biophys. J. 107, L9–L12 (2014).
Yan, P. et al. Palette of fluorinated voltage-sensitive hemicyanine dyes. Proc. Natl Acad. Sci. USA 109, 20443–20448 (2012).
Grenier, V., Walker, A. S. & Miller, E. W. A small-molecule photoactivatable optical sensor of transmembrane potential. J. Am. Chem. Soc. 137, 10894–10897 (2015).
Liu, P., Grenier, V., Hong, W., Muller, V. R. & Miller, E. W. Fluorogenic targeting of voltage-sensitive dyes to neurons. J. Am. Chem. Soc. 139, 17334–17340 (2017).
Grenier, V., Daws, B. R., Liu, P. & Miller, E. W. Spying on neuronal membrane potential with genetically targetable voltage indicators. J. Am. Chem. Soc. 141, 1349–1358 (2019).
Abdelfattah, A. S. et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699–704 (2019).
Uttamapinant, C. et al. Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew. Chem. Int. Ed. 51, 5852–5856 (2012).
Yao, J. Z. et al. Fluorophore targeting to cellular proteins via enzyme-mediated azide ligation and strain-promoted cycloaddition. J. Am. Chem. Soc. 134, 3720–3728 (2012).
Liu, D. S. et al. Diels–Alder cycloaddition for fluorophore targeting to specific proteins inside living cells. J. Am. Chem. Soc. 134, 792–795 (2012).
Xu, Y. et al. Hybrid indicators for fast and sensitive voltage imaging. Angew. Chem. Int. Ed. 57, 3949–3953 (2018).
Debets, M. F. et al. Bioconjugation with strained alkenes and alkynes. Acc. Chem. Res. 44, 805–815 (2011).
Lukinavicius, G. et al. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem. 5, 132–139 (2013).
Nikic, I. et al. Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew. Chem. Int. Ed. 53, 2245–2249 (2014).
Nikic, I., Kang, J. H., Girona, G. E., Aramburu, I. V. & Lemke, E. A. Labeling proteins on live mammalian cells using click chemistry. Nat. Protoc. 10, 780–791 (2015).
Beliu, G. et al. Bioorthogonal labeling with tetrazine-dyes for super-resolution microscopy. Commun. Biol. 2, 261 (2019).
Versteegen, R. M., Rossin, R., ten Hoeve, W., Janssen, H. M. & Robillard, M. S. Click to release: instantaneous doxorubicin elimination upon tetrazine ligation. Angew. Chem. Int. Ed. 52, 14112–14116 (2013).
Li, J., Jia, S. & Chen, P. R. Diels–Alder reaction-triggered bioorthogonal protein decaging in living cells. Nat. Chem. Biol. 10, 1003–1005 (2014).
Fan, X. et al. Optimized tetrazine derivatives for rapid bioorthogonal decaging in living cells. Angew. Chem. Int. Ed. 55, 14046–14050 (2016).
Li, J. & Chen, P. R. Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol. 12, 129–137 (2016).
Carlson, J. C. T., Mikula, H. & Weissleder, R. Unraveling tetrazine-triggered bioorthogonal elimination enables chemical tools for ultrafast release and universal cleavage. J. Am. Chem. Soc. 140, 3603–3612 (2018).
Kralj, J. M., Hochbaum, D. R., Douglass, A. D. & Cohen, A. E. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science 333, 345–348 (2011).
Kralj, J. M., Douglass, A. D., Hochbaum, D. R., Maclaurin, D. & Cohen, A. E. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods 9, 90–95 (2012).
Maclaurin, D., Venkatachalam, V., Lee, H. & Cohen, A. E. Mechanism of voltage-sensitive fluorescence in a microbial rhodopsin. Proc. Natl Acad. Sci. USA 110, 5939–5944 (2013).
Mujumdar, R. B., Ernst, L. A., Mujumdar, S. R., Lewis, C. J. & Waggoner, A. S. Cyanine dye labeling reagents: sulfoindocyanine succinimidyl esters. Bioconjug. Chem. 4, 105–111 (1993).
Bennett, M. V. & Zukin, R. S. Electrical coupling and neuronal synchronization in the mammalian brain. Neuron 41, 495–511 (2004).
Dhein, S. Gap junction channels in the cardiovascular system: pharmacological and physiological modulation. Trends Pharmacol. Sci. 19, 229–241 (1998).
Chow, B. Y. et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463, 98–102 (2010).
Kiskinis, E. et al. All-optical electrophysiology for high-throughput functional characterization of a human iPSC-derived motor neuron model of ALS. Stem Cell Rep. 10, 1991–2004 (2018).
Xu, Y. et al. Imaging neuronal activity with fast and sensitive red-shifted electrochromic FRET indicators. ACS Chem. Neurosci. 10, 4768–4775 (2019).
Uttamapinant, C., Sanchez, M. I., Liu, D. S., Yao, J. Z. & Ting, A. Y. Site-specific protein labeling using PRIME and chelation-assisted click chemistry. Nat. Protoc. 8, 1620–1634 (2013).
Acknowledgements
We acknowledge funding from the Ministry of Science and Technology (2018YFA0507600, 2017YFA0503600 and 2016YFA0501500), the National Natural Science Foundation of China (91753131, 32088101, 21673009, 21521003 and 21937001), the Natural Science Foundation of Beijing Municipality (5182011) and the Interdisciplinary Medicine Seed Fund of Peking University (BMU2017MC006). P.Z. was sponsored by a Li Ge-Zhao Ning Life Science Junior Research Fellowship, a Bayer Investigator Award and a National Thousand Young Talents Award. P.R.C. is the recipient of the XPLORER PRIZE from the Tencent Foundation. We thank Y. Li, H. Cheng, A. Cohen and L. Brown for providing plasmids. Z. Wu and Y. Li helped provide rat hippocampal neurons.
Author information
Authors and Affiliations
Contributions
P.Z. and P.R.C. conceived and supervised the project. S.L., C.L., Y.X. and L.P. performed all experiments, unless otherwise noted. C.L. and H.L. carried out chemical syntheses. X.Z. assisted with neuron culture. H.Z. assisted with purification of plasmids and enzymes. S.L., C.L., P.R.C. and P.Z. analysed data. S.L., C.L. and P.Z. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemistry thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Optimization of site-specific membrane protein labeling strategy.
a, Top: Probe incorporation mediated by enzyme (PRIME) coupled with copper-assisted alkyne-azide cycloaddition (CuAAC). Middle: PRIME coupled with strain-promoted alkyne-azide cycloaddition (SPAAC). Bottom: PRIME coupled with inverse-electron-demand Diels–Alder cycloaddition (IEDDA). b, LAP-Ace(D81N) domain structure. c, Representative confocal images of rat hippocampal neurons expressing LAP-Ace(D81N) and labeled with either DBCO-Cy3 (left) or Tz-Cy3 (right), scale bar = 20 μm. d, Label-to-background ratio of SPAAC and IEDDA conjugation methods. n = 10 and 10 cells, respectively. Error bars represent S.D. e, Simultaneous electrical and optical recording of stimulated action potentials from cultured neurons labeled with Cy3 via either SPAAC (left) or IEDDA (right). Cy3 traces were recorded from single trial measurements without temporal averaging. f,g, Cultured rat hippocampal neurons at DIV15-16 were labeled with 5 μM LplA and 100 μM 4-TCO for 30 min, before labeled with Tz-Cy3 via IEDDA at various probe concentrations and incubation times. (f) IEDDA labeling with 500 nM Tz-Cy3 for 1 to 15 min. n = 27 cells (1 min), 19 cells (2.5 min), 19 cells (5 min), 20 cells (10 min) and 22 cells (15 min). (g) IEDDA labeling for 10 min with 50 to 1000 nM Tz-Cy3. n = 13 cells (50 nM), 14 cells (100 nM), 18 cells (250 nM), 17 cells (500 nM) and 26 cells (1000 nM). Whole cell fluorescence intensities were measured on an inverted fluorescence microscope and normalized to either 1 min or 50 nM. Error bars represent s.e.m.
Extended Data Fig. 2 Membrane trafficking and photocurrents of HVIs.
a, LAP-Ace(D81X)-mOrange2 domain structure. b, Representative confocal images of cultured rat hippocampal neurons expressing to Ace(D81N) and Ace(D81C) mutants labeled with Tz-Cy5. Top: mOrange2 images showing the expression of Ace fusion. Middle: Cy5 images showing IEDDA fluorescence label. Bottom: overlay of mOrange2 and Cy5 channels. Scale bars = 20 μm. Photocurrent measurements were performed on DIV12–14. Cells expressing HVI were labeled with fluorophores and voltage-clamped at -70 mV via a patch pipet. c, Representative electrical traces of photocurrents in cultured neurons expressing HVI under a 300-ms light illumination pulse at laser wavelengths of 488 nm (3.1 W/cm2, n = 4 cells), 532 nm (4.9 W/cm2, n = 5 cells), 594 nm (3.8 W/cm2, n = 8 cells) and 637 nm (0.48 W/cm2, n = 5 cells), respectively. d,e, The steady-state photocurrents (d) and peak photocurrents (e) of HVIs and VARNAM (561 nm, 4.8 W/cm2) in response to different laser wavelengths, measured in cultured rat hippocampal neurons. Error bars represent s.e.m.
Extended Data Fig. 3 Mapping electrical connectivity of cultured HEK293T cells.
a, A representative 333 μm × 333 μm wide-field fluorescence image of HEK293T cells expressing HVI-Cy3 (top). An overlay of Cy3 fluorescence with DIC image is shown at the bottom. Scale bars = 20 μm. b, F-V response curves of cells indicated in a. The membrane potential of cell #1 (red) was controlled via whole-cell voltage clamp at 1000 Hz, and ramped between -100 mV to +50 mV. Fluorescence images of the whole field were acquired at a camera frame rate of 10 Hz. The fluorescence response had been normalized to cell #1. c, Single exponential fitting of normalized response versus the distance to the clamped cell (grey dashed line). The decay length constant is calculated to be 99.2 μm. d, Fluorescence response to step-wise voltage changes, similar to b, except that the membrane potential was stepped between -70 mV and 30 mV, while cells in a 166 μm × 166 μm field of view were imaged at 200 Hz camera frame rate. The fluorescence signals had been normalized to the fluorescence at Vm = -70 mV and 30 mV. e, A zoomed-in view of the shaded region in d, showing the normalized fluorescence response during a depolarization voltage step. The time point at step voltage change is defined as t = 0.
Extended Data Fig. 4 Fluorescence images and photostability of a panel of voltage indicators.
a-f, Left: representative confocal images of rat hippocampal neurons expressing HVI-488 (a), HVI-Cy3 (b), HVI-594 (c), HVI-Cy5 (d), Ace2N-mNeon (e), and VARNAM (f). Scale bars = 20 μm. Right: single trial recordings membrane voltage and fluorescence response of these indicators at 484 Hz camera frame rate. g, Representative photobleaching curves are shown for VARNAM, Ace2N-mNeon, and HVIs, measured in HEK293T cells. Photobleaching constants are summarized in Supplementary Table 6. Image sampling rate is 20 Hz. Fluorescence intensities were normalized to the initial value at t = 0.
Extended Data Fig. 5 Wide-field voltage imaging of multiple neurons with HVI-Cy3.
a, A representative 452 μm × 161 μm wide-field image of cultured rat hippocampal neurons expressing HVI-Cy3 (left). An overlay with DIC image is shown on the right. Scale bars = 50 μm. b, Wide-field voltage imaging of HVI-Cy3 at 400 Hz over 10 min under ~2 W/cm2 illumination with 20× NA1.05 dry objective lens. The fluorescence traces of four individual neurons are shown during the time window of 0-100 s, 300-400 s, and 400-500 s. At t = 300 s, 5 mM KCl was added to stimulate AP firing. c, A zoom-in view of the shaded region in b.
Extended Data Fig. 6 Simultaneous dual-color imaging of pH or extracellular glutamate and membrane voltage in neurons.
a, Representative confocal images of rat hippocampal neurons co-expressing a membrane-anchored pHluorin-CAAX (left) and HVI-Cy5 voltage indicator (middle). The overlay image of pHluorin and Cy5 is shown on the right. Scale bar = 20 μm. b, Dual-color imaging of pHluorin (green trace, 488 nm, 2.5 W/cm2) and HVI-Cy5 (magenta trace, 637 nm, 0.9 W/cm2) at 484 Hz for 100 s. The signals of pHluorin and Cy5 had been corrected for photobleaching. c, A zoom-in view of the shaded region in b. Individual APs are marked with asterisks. d, Glutamate (top) and membrane voltage (bottom) traces were monitored with SF-iGluSnFR and HVI-Cy5, respectively, at a camera frame rate of 484 Hz. e, A zoom-in view of the shaded region in d. Cy5 traces have been corrected for photobleaching. Individual APs are marked with asterisks. f, A linear regression of the glutamate signal versus the number of recorded AP spikes in clusters. n = 30, 21, 8, 6, 7, 2, 6, and 3 cells, respectively. Error bars represent s.e.m. g, Heatmap images at 8-by-8 binning showing the ΔF/F0 and baseline fluorescence (F0) of SF-iGluSnFR during the time window indicated by the shaded region in d.
Extended Data Fig. 7 All-optical electrophysiology with HVI-Cy5 with CheRiff.
a, Representative confocal images of rat hippocampal neurons co-expressing CheRiff-EGFP and HVI-Cy5. Scale bar = 20 μm. b, Neurons were stimulated to fire individual APs upon illumination with 2-ms blue-light pulses. Membrane voltage and fluorescence responses were recorded simultaneously via patch pipette and camera at a frame of 821 Hz, respectively. c, A zoom-in view of the shaded region in b. d, Neurons were stimulated to fire AP spike trains via 250-ms blue-light illumination. Image sampling rate was 464 Hz.
Extended Data Fig. 8 Characterization of optical and optogenetic crosstalk among HVI-Cy5, CheRiff, and R-GECO1 or Mac.
a, Overlay of the absorption spectrum of CheRiff (dashed curve), and the excitation spectra of R-GECO1 (yellow) and Cy5 (red). Blue, yellow and red bars represent 488 nm, 561 nm and 637 nm laser lines respectively. b, Steady-state photocurrent generated by 488 nm (0.03 W/cm2, -371 ± 35 pA, n = 15 cells), 561 nm (0.10 W/cm2, -18.0 ± 3.4 pA, n = 6 cells) or 637 nm (0.36 W/cm2, 0.0 ± 0.1 pA, n = 10 cells) in CheRiff-expressing neurons. c, The emission spectra of R-GECO1 (orange) and Cy5 (red). The emission filters used in this study are indicated as shaded regions. d, Neurons expressing R-GECO1 were illuminated by 488 nm (0.03 W/cm2), 561 nm (0.10 W/cm2) and 637 nm (0.36 W/cm2), respectively. The optical crosstalk (measured with the emission filter 600/50) was normalized to 561 nm illumination (n = 6 neurons). Error bars represent s.e.m. e, Overlay of the action spectra of Mac45 (grey dashed curve) and CheRiff10 (orange dashed curve) and the excitation spectrum of Cy5 (red). Blue, yellow and red bars represent 488 nm, 561 nm and 637 nm laser lines, respectively. f, Representative HVI-Cy5 fluorescence trace of cultured neuron before and after 4.0% optical crosstalk correction (see below). Blue bar: 2153-ms 488 nm light illumination at 0.03 W/cm2. Yellow bar: 538-ms 561 nm light illumination at 0.48 W/cm2. Cy5 fluorescence was recorded under continuous 637 nm illumination at 0.24 W/cm2. g, Whole-cell fluorescence changes of neurons expressing HVI-Cy5 under 488 nm and/or 561 nm laser illuminations. 561 nm illumination caused a 4.0% increase in Cy5 fluorescence, whereas 488 nm laser illumination had negligible effect (n = 7 neurons). h, Representative photocurrent traces over 30 repeated trails. i,j, Steady-state photocurrent (i, n = 5 cells) and membrane depolarization (j, n = 4 cells) generated by 488 nm (0.06 W/cm2), 561 nm (0.48 W/cm2) or 637 nm (0.24 W/cm2) illumination of neurons expressing Mac-P2A-CheRiff. k, Simultaneous electrical and optical recordings of APs from a neuron co-expressing Mac-P2A-CheRiff and HVI-Cy5. Blue-light stimulation caused AP firing, which was interrupted by yellow light pulses. Error bars represent s.e.m.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and Tables 1–10.
Rights and permissions
About this article
Cite this article
Liu, S., Lin, C., Xu, Y. et al. A far-red hybrid voltage indicator enabled by bioorthogonal engineering of rhodopsin on live neurons. Nat. Chem. 13, 472–479 (2021). https://doi.org/10.1038/s41557-021-00641-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-021-00641-1