Abstract
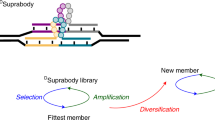
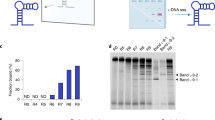
The physicochemical properties of nucleic acids are dominated by their highly charged phosphodiester backbone chemistry. This polyelectrolyte structure decouples information content (base sequence) from bulk properties, such as solubility, and has been proposed as a defining trait of all informational polymers. However, this conjecture has not been tested experimentally. Here, we describe the encoded synthesis of a genetic polymer with an uncharged backbone chemistry: alkyl phosphonate nucleic acids (phNAs) in which the canonical, negatively charged phosphodiester is replaced by an uncharged P-alkyl phosphonodiester backbone. Using synthetic chemistry and polymerase engineering, we describe the enzymatic, DNA-templated synthesis of P-methyl and P-ethyl phNAs, and the directed evolution of specific streptavidin-binding phNA aptamer ligands directly from random-sequence mixed P-methyl/P-ethyl phNA repertoires. Our results establish an example of the DNA-templated enzymatic synthesis and evolution of an uncharged genetic polymer and provide a foundational methodology for their exploration as a source of novel functional molecules.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information files. The molecular modelling data and related settings for computations that support the findings of this study are available in the Zenodo database (https://zenodo.org/) with the following record 2579703 (https://doi.org/10.5281/zenodo.2579703).
References
Westheimer, F. H. Why nature chose phosphates. Science 235, 1173–1178 (1987).
Benner, S. A. Understanding nucleic acids using synthetic chemistry. Acc. Chem. Res. 37, 784–797 (2004).
Benner, S. A. & Hutter, D. Phosphates, DNA, and the search for nonterrean life: a second generation model for genetic molecules. Bioorg. Chem. 30, 62–80 (2002).
Malyshev, D. A. & Romesberg, F. E. The expanded genetic alphabet. Angew. Chem. Int. Ed. 54, 11930–11944 (2015).
Pinheiro, V. B. & Holliger, P. The XNA world: progress towards replication and evolution of synthetic genetic polymers. Curr. Opin. Chem. Biol. 16, 245–252 (2012).
Pinheiro, V. B. et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 (2012).
Taylor, A. I. et al. Catalysts from synthetic genetic polymers. Nature 518, 427–430 (2015).
Malyshev, D. A. et al. A semi-synthetic organism with an expanded genetic alphabet. Nature 509, 385–388 (2014).
Liu, C. et al. Phosphonomethyl oligonucleotides as backbone-modified artificial genetic polymers. J. Am. Chem. Soc. 140, 6690–6699 (2018).
Zhang, S. L., Blain, J. C., Zielinska, D., Gryaznov, S. M. & Szostak, J. W. Fast and accurate nonenzymatic copying of an RNA-like synthetic genetic polymer. Proc. Natl Acad. Sci. USA 110, 17732–17737 (2013).
Ghadessy, F. J. et al. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat. Biotechnol. 22, 755–759 (2004).
Shaw, B. R. et al. Reading, writing, and modulating genetic information with boranophosphate mimics of nucleotides, DNA, and RNA. Ann. NY Acad. Sci. 1002, 12–29 (2003).
King, D. J., Ventura, D. A., Brasier, A. R. & Gorenstein, D. G. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry 37, 16489–16493 (1998).
Meng, M. & Ducho, C. Oligonucleotide analogues with cationic backbone linkages. Beilstein J. Org. Chem. 14, 1293–1308 (2018).
Nielsen, P. E. DNA analogues with nonphosphodiester backbones. Annu. Rev. Biophys. Biomol. Struct. 24, 167–183 (1995).
Steinbeck, C. & Richert, C. The role of ionic backbones in RNA structure: an unusually stable non-Watson–Crick duplex of a nonionic analog in an apolar medium. J. Am. Chem. Soc. 120, 11576–11580 (1998).
Micklefield, J. Backbone modification of nucleic acids: synthesis, structure and therapeutic applications. Curr. Med. Chem. 8, 1157–1179 (2001).
Summerton, J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta 1489, 141–158 (1999).
Nielsen, P. E. & Egholm, M. An introduction to peptide nucleic acid. Curr. Issues Mol. Biol. 1, 89–104 (1999).
Dineva, M. A., Chakurov, S., Bratovanova, E. K., Devedjiev, I. & Petkov, D. D. Complete template-directed enzymatic synthesis of a potential antisense DNA containing 42 methylphosphonodiester bonds. Bioorgan. Med. Chem. 1, 411–414 (1993).
Higuchi, H., Endo, T. & Kaji, A. Enzymic synthesis of oligonucleotides containing methylphosphonate internucleotide linkages. Biochemistry 29, 8747–8753 (1990).
Brudno, Y., Birnbaum, M. E., Kleiner, R. E. & Liu, D. R. An in vitro translation, selection and amplification system for peptide nucleic acids. Nat. Chem. Biol. 6, 148–155 (2010).
Murakami, A., Blake, K. R. & Miller, P. S. Characterization of sequence-specific oligodeoxyribonucleoside methylphosphonates and their interaction with rabbit globin mRNA. Biochemistry 24, 4041–4046 (1985).
Arzumanov, A. A. & Dyatkina, N. B. An alternative route for preparation of α-methylphosphonyl-β,γ-diphosphates of thymidine derivatives. Nucleos. Nucleot. 13, 1031–1037 (1994).
Burgers, P. M. J. & Eckstein, F. Stereochemistry of internucleotide bond formation by polynucleotide phosphorylase from Micrococcus luteus. Biochemistry 18, 450–454 (1979).
Xia, S. & Konigsberg, W. H. Mispairs with Watson–Crick base-pair geometry observed in ternary complexes of an RB69 DNA polymerase variant. Protein Sci. 23, 508–513 (2014).
Genna, V., Gaspari, R., Dal Peraro, M. & De Vivo, M. Cooperative motion of a key positively charged residue and metal ions for DNA replication catalyzed by human DNA polymerase-η. Nucleic Acids Res. 44, 2827–2836 (2016).
Genna, V., Donati, E. & De Vivo, M. The catalytic mechanism of DNA and RNA polymerases. ACS Catal. 8, 11103–11118 (2018).
Genna, V., Carloni, P. & De Vivo, M. A strategically located Arg/Lys residue promotes correct base paring during nucleic acid biosynthesis in polymerases. J. Am. Chem. Soc. 140, 3312–3321 (2018).
Genna, V., Colombo, M., De Vivo, M. & Marcia, M. Second-shell basic residues expand the two-metal-ion architecture of DNA and RNA processing enzymes. Structure 26, 40–50.e2 (2018).
Cozens, C., Pinheiro, V. B., Vaisman, A., Woodgate, R. & Holliger, P. A short adaptive path from DNA to RNA polymerases. Proc. Natl Acad. Sci. USA 109, 8067–8072 (2012).
Wynne, S. A., Pinheiro, V. B., Holliger, P. & Leslie, A. G. Structures of an apo and a binary complex of an evolved archeal B family DNA polymerase capable of synthesising highly Cy-dye labelled DNA. PLoS ONE 8, e70892 (2013).
Bergen, K., Betz, K., Welte, W., Diederichs, K. & Marx, A. Structures of KOD and 9°N DNA polymerases complexed with primer template duplex. ChemBioChem 14, 1058–1062 (2013).
Genna, V., Vidossich, P., Ippoliti, E., Carloni, P. & De Vivo, M. A self-activated mechanism for nucleic acid polymerization catalyzed by DNA/RNA polymerases. J. Am. Chem. Soc. 138, 14592–14598 (2016).
Nakamura, T., Zhao, Y., Yamagata, Y., Hua, Y. J. & Yang, W. Watching DNA polymerase η make a phosphodiester bond. Nature 487, 196–201 (2012).
Pinheiro, V. B., Loakes, D. & Holliger, P. Synthetic polymers and their potential as genetic materials. BioEssays 35, 113–122 (2013).
Dunn, M. R. & Chaput, J. C. Reverse transcription of threose nucleic acid by a naturally occurring DNA polymerase. ChemBioChem 17, 1804–1808 (2016).
Thiviyanathan, V. et al. Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and I stereochemistry. Biochemistry 41, 827–238 (2002).
Vyazovkina, E. V. et al. Synthesis of specific diastereomers of a DNA methylphosphonate heptamer, d(CpCpApApApCpA), and stability of base pairing with the normal DNA octamer d(TpGpTpTpTpGpGpC). Nucleic Acids Res. 22, 2404–2409 (1994).
Tsai, C. H., Chen, J. & Szostak, J. W. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc. Natl Acad. Sci. USA 104, 14598–14603 (2007).
Burmeister, P. E. et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 12, 25–33 (2005).
Alves Ferreira-Bravo, I., Cozens, C., Holliger, P. & DeStefano, J. J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 43, 9587–9599 (2015).
Yu, H., Zhang, S. & Chaput, J. C. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 4, 183–187 (2012).
Rangel, A. E., Chen, Z., Ayele, T. M. & Heemstra, J. M. In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res. 46, 8057–8068 (2018).
Lee, E. J., Lim, H. K., Cho, Y. S. & Hah, S. S. Peptide nucleic acids are an additional class of aptamers. RSC Adv. 3, 5828–5831 (2013).
Ichida, J. K. et al. An in vitro selection system for TNA. J. Am. Chem. Soc. 127, 2802–2803 (2005).
Bing, T., Yang, X. J., Mei, H. C., Cao, Z. H. & Shangguan, D. H. Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorg. Med. Chem. 18, 1798–1805 (2010).
Weber, P. C., Ohlendorf, D. H., Wendoloski, J. J. & Salemme, F. R. Structural origins of high-affinity biotin binding to streptavidin. Science 243, 85–88 (1989).
Freitag, S., LeTrong, I., Klumb, L., Stayton, P. S. & Stenkamp, R. E. Structural studies of the streptavidin binding loop. Protein Sci. 6, 1157–1166 (1997).
Houlihan, G., Arangundy-Franklin, S. & Holliger, P. Engineering and application of polymerases for synthetic genetics. Curr. Opin. Biotechnol. 48, 168–179 (2017).
Krishna, H. & Caruthers, M. H. Alkynyl phosphonate DNA: a versatile ‘click’able backbone for DNA-based biological applications. J. Am. Chem. Soc. 134, 11618–11631 (2012).
Acknowledgements
This work was supported by Trinity College Cambridge (S.A.,-F.), by the Medical Research Council (S.A.-F. A.I.T., S.P.-C., P.H., program no. MC_U105178804), by the Biotechnology and Biological Sciences Research Council (B.T.P., BBSRC grant no BB/N01023x/1), by the NICHD/ NIH Intramural Research Program (A.V. and R.W.) and by a European Molecular Biology Organization (EMBO) Long-Term Fellowship (V.G., ALTF 103-2018).
Author information
Authors and Affiliations
Contributions
S.A.-F. and P.H. conceived and designed the experiments. S.A.-F. performed all the experiments except the SPR measurements (A.T. and B.T.P.), MS (S.P.-C.) and steady-state kinetics (A.V. and R.W.) and Modelling and MD simulations (V.G. and M.O.). All the authors discussed the results, and jointly wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary figures, Supplementary tables and Supplementary references.
Rights and permissions
About this article
Cite this article
Arangundy-Franklin, S., Taylor, A.I., Porebski, B.T. et al. A synthetic genetic polymer with an uncharged backbone chemistry based on alkyl phosphonate nucleic acids. Nat. Chem. 11, 533–542 (2019). https://doi.org/10.1038/s41557-019-0255-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-019-0255-4
This article is cited by
-
Accessible light-controlled knockdown of cell-free protein synthesis using phosphorothioate-caged antisense oligonucleotides
Communications Chemistry (2023)
-
An RNA-cleaving threose nucleic acid enzyme capable of single point mutation discrimination
Nature Chemistry (2022)
-
Evaluation of 3′-phosphate as a transient protecting group for controlled enzymatic synthesis of DNA and XNA oligonucleotides
Communications Chemistry (2022)