Abstract
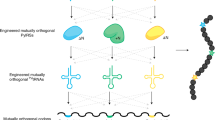
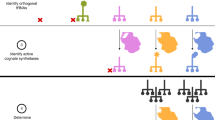
Genetically encoding distinct non-canonical amino acids (ncAAs) into proteins synthesized in cells requires mutually orthogonal aminoacyl-tRNA synthetase (aaRS)/tRNA pairs. The pyrrolysyl-tRNA synthetase/PyltRNA pair from Methanosarcina mazei (Mm) has been engineered to incorporate diverse ncAAs and is commonly considered an ideal pair for genetic code expansion. However, finding new aaRS/tRNA pairs that share the advantages of the MmPylRS/MmPyltRNA pair and are orthogonal to both endogenous aaRS/tRNA pairs and the MmPylRS/MmPyltRNA pair has proved challenging. Here we demonstrate that several ΔNPylRS/PyltRNACUA pairs, in which PylRS lacks an N-terminal domain, are active, orthogonal and efficiently incorporate ncAAs in Escherichia coli. We create new PylRS/PyltRNA pairs that are mutually orthogonal to the MmPylRS/MmPyltRNA pair and show that transplanting mutations that reprogram the ncAA specificity of MmPylRS into the new PylRS reprograms its substrate specificity. Finally, we show that distinct PylRS/PyltRNA-derived pairs can function in the same cell, decode distinct codons and incorporate distinct ncAAs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wang, K. et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat. Chem. 6, 393–403 (2014).
Sachdeva, A., Wang, K., Elliott, T. & Chin, J. W. Concerted, rapid, quantitative, and site-specific dual labeling of proteins. J. Am. Chem. Soc. 136, 7785–7788 (2014).
Chatterjee, A., Sun, S. B., Furman, J. L., Xiao, H. & Schultz, P. G. A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 52, 1828–1837 (2013).
Neumann, H., Wang, K., Davis, L., Garcia-Alai, M. & Chin, J. W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444 (2010).
Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017).
Neumann, H., Slusarczyk, A. L. & Chin, J. W. De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA pairs. J. Am. Chem. Soc. 132, 2142–2144 (2010).
Chatterjee, A., Xiao, H. & Schultz, P. G. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc. Natl Acad. Sci. USA 109, 14841–14846 (2012).
Anderson, J. C. et al. An expanded genetic code with a functional quadruplet codon. Proc. Natl Acad. Sci. USA 101, 7566–7571 (2004).
Chin, J. W. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 83, 379–408 (2014).
Ambrogelly, A. et al. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl Acad. Sci. USA 104, 3141–3146 (2007).
Elliott, T. S. et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol. 32, 465–472 (2014).
Dumas, A., Lercher, L., Spicer, C. D. & Davis, B. G. Designing logical codon reassignment—expanding the chemistry in biology. Chem. Sci. 6, 50–69 (2015).
Wang, K. et al. Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64 (2016).
Ostrov, N. et al. Design, synthesis, and testing toward a 57-codon genome. Science 353, 819–822 (2016).
Zhang, Y. et al. A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551, 644–647 (2017).
Jiang, R. & Krzycki, J. A. PylSn and the homologous N-terminal domain of pyrrolysyl-tRNA synthetase bind the tRNA that is essential for the genetic encoding of pyrrolysine. J. Biol. Chem. 287, 32738–32746 (2012).
Suzuki, T. et al. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 13, 1261–1266 (2017).
Kavran, J. M. et al. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl Acad. Sci. USA 104, 11268–11273 (2007).
Herring, S. et al. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 581, 3197–3203 (2007).
Sollinger, A. Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences. FEMS Microbiol. Ecol. 92, pii: fiv149 (2016).
Borrel, G. et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genom. 15, 679 (2014).
Virdee, S., Ye, Y., Nguyen, D. P., Komander, D. & Chin, J. W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 6, 750–757 (2010).
Yanagisawa, T. et al. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode Nɛ-(o-Azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol. 15, 1187–1197 (2008).
Neumann, H., Peak-Chew, S. Y. & Chin, J. W. Genetically encoding Nε-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234 (2008).
Zhang, M. S. et al. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing. Nat. Methods 14, 729–736 (2017).
Borrmann, A. et al. Genetic encoding of a bicyclo[6.1.0]nonyne-charged amino acid enables fast cellular protein imaging by metal-free ligation. ChemBioChem 13, 2094–2099 (2012).
Lang, K. et al. Genetic encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels–Alder reactions. J. Am. Chem. Soc. 134, 10317–10320 (2012).
Acknowledgements
This work was supported by the Medical Research Council, UK (MC_U105181009 and MC_UP_A024_1008), BBSRC (BB/M000842/1, for automation) and an ERC Advanced Grant (SGCR), all to J.W.C. J.C.W.W. was supported by the EPSRC Nanoscience and Nanotechnology CDT at Cambridge University.
Author information
Authors and Affiliations
Contributions
J.C.W.W. performed all experiments. J.C.W.W and J.W.C. conceived and designed experiments, analysed the data, and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
A single Supplementary Information file contains the Methods and Supplementary Figures
Rights and permissions
About this article
Cite this article
Willis, J.C.W., Chin, J.W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nature Chem 10, 831–837 (2018). https://doi.org/10.1038/s41557-018-0052-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-018-0052-5
This article is cited by
-
Deciphering functional roles of protein succinylation and glutarylation using genetic code expansion
Nature Chemistry (2024)
-
Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-α-amino acids in vitro and in vivo
Nature Chemistry (2023)
-
Five mutually orthogonal aaRS–tRNA pairs for genetic code expansion
Nature Chemistry (2023)
-
Quintuply orthogonal pyrrolysyl-tRNA synthetase/tRNAPyl pairs
Nature Chemistry (2023)
-
Light-activated tetrazines enable precision live-cell bioorthogonal chemistry
Nature Chemistry (2022)