Abstract
Protein trafficking requires coat complexes that couple recognition of sorting motifs in transmembrane cargoes with biogenesis of transport carriers. The mechanisms of cargo transport through the endosomal network are poorly understood. Here, we identify a sorting motif for endosomal recycling of cargoes, including the cation-independent mannose-6-phosphate receptor and semaphorin 4C, by the membrane tubulating BAR domain-containing sorting nexins SNX5 and SNX6. Crystal structures establish that this motif folds into a β-hairpin, which binds a site in the SNX5/SNX6 phox homology domains. Over sixty cargoes share this motif and require SNX5/SNX6 for their recycling. These include cargoes involved in neuronal migration and a Drosophila snx6 mutant displays defects in axonal guidance. These studies identify a sorting motif and provide molecular insight into an evolutionary conserved coat complex, the ‘Endosomal SNX–BAR sorting complex for promoting exit 1’ (ESCPE-1), which couples sorting motif recognition to the BAR-domain-mediated biogenesis of cargo-enriched tubulo-vesicular transport carriers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The coordinates and structure factors for the SNX5 PX domain in complex with the CI-MPR sorting signal have been deposited at the PDB with accession codes 6N5X (form 1) and 6N5Y (form 2). The coordinates and structure factors for the SNX5 PX domain in complex with the SEMA4C sorting signal have been deposited at the PDB with the accession code 6N5Z. Supplementary Table 3 contains all of the raw data from the proteomic analysis shown in Fig. 7b,f. All of the other relevant raw data related to this study are available from the corresponding authors on request. The mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD014927. The source data for Fig. 8a,d have been provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
Goldenring, J. R. Recycling endosomes. Curr. Opin. Cell Biol. 35, 117–122 (2015).
Cullen, P. J. & Steinberg, F. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19, 679–696 (2018).
Behnia, R. & Munro, S. Organelle identity and the signposts for membrane traffic. Nature 438, 597–604 (2005).
McNally, K. E. & Cullen, P. J. Endosomal retrieval of cargo: retromer is not alone. Trends Cell Biol. 28, 807–822 (2018).
Phillips-Krawczak, C. A. et al. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol. Biol. Cell 26, 91–103 (2015).
Bartuzi, P. et al. CCC- and WASH-mediated endosomal sorting of the LDLR is required for normal clearance of circulating LDL. Nat. Commun. 7, 10961 (2016).
Meyer, C. et al. μ1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 19, 2193–2203 (2000).
Seaman, M. N. J., McCaffery, J. M. & Emr, S. D. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142, 665–681 (1998).
Seaman, M. N. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165, 111–122 (2004).
Arighi, C. N., Hartnell, L. M., Aguilar, R. C., Haft, C. R. & Bonifacino, J. S. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165, 123–133 (2004).
Strochlic, T. I., Setty, T. G., Sitaram, A. & Burd, C. G. Grd19/Sns3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 177, 115–125 (2007).
Harterink, M. et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13, 914–923 (2011).
Temkin, P. et al. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13, 715–721 (2011).
Steinberg, F. et al. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15, 461–471 (2013).
Lucas, M. et al. Structural mechanism for cargo recognition by the retromer complex. Cell 167, 1623–1635 (2016).
McNally, K. E. et al. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol. 19, 1214–1225 (2017).
Purushothaman, L. K. & Ungermann, C. Cargo induces retromer-mediated membrane remodelling on membranes. Mol. Biol. Cell 29, 2709–2719 (2018).
Kovtun, O. et al. Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature 561, 561–564 (2018).
Carlton, J. et al. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr. Biol. 14, 1791–1800 (2004).
Wassmer, T. et al. A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. J. Cell Sci. 120, 45–54 (2007).
Wassmer, T. et al. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport with carrier recognition by the trans-Golgi network. Dev. Cell 17, 110–122 (2009).
van Weering, J. R. et al. Molecular basis for SNX–BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 31, 4466–4480 (2012).
Simonetti, B., Danson, C. M., Heesom, K. J. & Cullen, P. J. Sequence-dependent cargo recognition by SNX–BARs mediates retromer-independent transport of CI-MPR. J. Cell Biol. 216, 3695–3712 (2017).
Kvainickas, A. et al. Cargo-selective SNX–BAR proteins mediate retromer trimer independent retrograde transport. J. Cell Biol. 216, 3677–3693 (2017).
Teasdale, R. D., Loci, D., Houghton, F., Karlsson, L. & Gleeson, P. A. A large family of endosome-localised proteins related to sorting nexin 1. Biochem. J. 358, 7–16 (2001).
Elwell, C. A. et al. Chlamydia interfere with an interaction between the mannose-6-phosphate receptor and sorting nexins to counteract host restriction. eLife 6, e22709 (2017).
Paul, B. et al. Structural basis for the hijacking of endosomal sorting nexin proteins by Chlamydia trachomatis. eLife 6, e22311 (2017).
Sun, Q. et al. Structural and functional insights into sorting nexin 5/6 interaction with bacterial effector IncE. Signal Transduct. Target. Ther. 2, 17030 (2017).
Seaman, M. N. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J. Cell Sci. 120, 2378–2389 (2007).
Coutinho, M. F., Prata, M. J. & Alves, S. Mannose-6-phosphate pathway: a review on its role in lysosomal function and dysfunction. Mol. Genet. Metab. 105, 542–550 (2012).
Itai, N. et al. The phosphorylation of sorting nexin 5 at serine 226 regulates retrograde transport and macropinocytosis. PLoS One 13, e0207205 (2018).
Chandra, M. et al. Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat. Commun. 10, 1528 (2019).
Blockus, H. & Chedotal, A. Slit-Robo signaling. Development 143, 3037–3044 (2016).
Deng, H. X. et al. Identification of TMEM230 mutations in familial Parkinson’s disease. Nat. Genet. 48, 733–739 (2016).
Wojciech, S. et al. The orphan GPR50 receptor promotes constitutive TGFβ receptor signaling and protects against cancer development. Nat. Commun. 9, 1216 (2018).
Fan, X., Labrador, J. P., Hing, H. & Bashaw, G. J. Slit stimulation recruits Dock and Pak to the roundabout receptor and increases rac activity to regulate axon repulsion at the CNS midline. Neuron 40, 113–127 (2003).
Kidd, T., Bland, K. S. & Goodman, C. S. Slit is the midline repellent for the Robo receptor in Drosophila. Cell 96, 785–794 (1999).
Coleman, H. A., Labrador, J. P., Chance, R. K. & Bashaw, G. J. The Adam family metalloprotease Kuzbanian regulates the cleavage of the roundabout receptor to control axon repulsion at the midline. Development 137, 2417–2426 (2010).
Hernandez-Fleming, M., Rohrbach, E. & Bashaw, G. J. Sema-1a reverse signaling promotes midline crossing in response to secreted Semaphorins. Cell Rep. 18, 174–184 (2017).
Klumperman, J. & Raposo, G. The complex ultrastrutcure of the endolysosomal system. Cold Spring Harb. Perspect. Biol. 6, a016857 (2014).
Bright, N. A., Davis, L. J. & Luzio, J. P. Endolysosomes are the principal intracellular sites of acid hydrolase activity. Curr. Biol. 26, 2233–2245 (2016).
Mari, M. et al. SNX1 defines an early endosomal recycling exit for sortilin and mannose 6-phosphate receptors. Traffic 9, 380–393 (2008).
Johannes, L. & Wunder, C. Retrograde transport: two (or more) roads diverged in an endosomal tree? Traffic 12, 956–962 (2011).
Seaman, M. N. J. Retromer and the cation-independent mannose 6-phosphate receptor—time for a trial separation? Traffic 19, 150–152 (2018).
Carroll, K. S. et al. Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science 292, 1373–1376 (2001).
Wan, L. et al. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localisation. Cell 94, 205–216 (1998).
Robinson, M. S., Sahlender, D. A. & Foster, S. D. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev. Cell 18, 324–331 (2010).
Hirst, J. et al. Interaction between AP-5 and the hereditary spastic paraplegia proteins SPG11 and SPG15. Mol. Biol. Cell 24, 2558–2569 (2013).
Strutt, H. et al. Retromer controls planar polarity protein levels and asymmetric localization at intercellular junctions. Curr. Biol. 29, 484–491 (2019).
Burd, C. & Cullen, P. J. Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 6, a016774 (2014).
Gallon, M.et al. A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc. Natl Acad. Sci. USA 111, E3604–13 (2014).
Gomez, T. S. & Billadeau, D. D. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711 (2009).
Harbour, M. E., Breusegem, S. Y. & Seaman, M. N. Recruitment of the endosomal WASH complex is mediated by the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. Biochem. J. 442, 209–220 (2012).
Jia, D., Gomez, T. S., Billadeau, D. D. & Rosen, M. K. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol. Biol. Cell 23, 2352–2361 (2012).
McGough, I. J. et al. Identification of molecular heterogeneity in SNX27-retromer-mediated endosome-to-plasma membrane recycling. J. Cell Sci. 127, 4940–4953 (2014).
Kvainickas, A. et al. Retromer- and WASH-dependent sorting of nutrient transporters requires a multivalent interaction network with ANKRD50. J. Cell Sci. 130, 382–395 (2017).
Cui, Y. et al. Retromer has a selective function in cargo sorting via endosome transport carriers. J. Cell Biol. 218, 615–631 (2019).
Hirst, J. et al. Distinct and overlapping roles for AP-1 and GGAs revealed by the ‘knocksideways’ system. Curr. Biol. 22, 1711–1716 (2012).
Hirst, J., Itzhak, D. N., Antrobus, R., Borner, G. H. H. & Robinson, M. S. Role of the AP-5 adaptor protein complex in late endosome-to-Golgi retrieval. PLoS Biol. 16, e2004411 (2018).
Varandas, K. C., Irannejad, R. & von Zastrow, M. Retromer endosome exit domains serve multiple trafficking destinations and regulate local G protein activation by GPCRs. Curr. Biol. 26, 3129–3142 (2016).
Shafaq-Zadah, M. et al. Presistent cell migration and adhesion rely on retrograde transport of β1-integrin. Nat. Cell Biol. 18, 54–64 (2016).
Mirrashidi, K. M. et al. Global mapping of the Inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe 18, 109–121 (2015).
Subbarayal, P. et al. EphrinA2 receptor (EphA2) is an invasion and intracellular signalling receptor for Chlamydia trachomatis. PLoS Pathog. 11, e1004846 (2015).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011).
Acknowledgements
This work was supported by the MRC (grant nos MR/L007363/1 and MR/P018807/1), the Wellcome Trust (grant no. 104568/Z/14/2), the Lister Institute of Preventive Medicine to P.J.C. and the Wellcome Trust PhD Studentship for the Dynamic Cell Biology programme (grant no. 083474) to B.S. B.M.C. is supported by an NHMRC Senior Research Fellowship (grant no. APP1136021) and the NHMRC (grant no. APP1099114), the Australian Research Council (grant no. DP160101743) and the Bright Focus Foundation (grant no. A2018627S). G.J.B. is supported by the National Institutes of Health (grant no. R35 NS097340). We thank the University of Queensland Remote Operation Crystallisation and X-ray facility and the staff for their support with the crystallization experiments, the staff of the Australian Synchrotron for assistance with X-ray diffraction data collection and the University of Bristol Wolfson Bioimaging Facility.
Author information
Authors and Affiliations
Contributions
Initial conceptualization: B.S., F.S., B.M.C. and P.J.C. Evolution of conceptualization: B.S., B.P., S.W., F.S., G.J.B., B.M.C. and P.J.C. Formal analysis: K.J.H. Investigation: B.S., B.P., K.C., S.W., M.G., F.S. and K.J.H. Writing of the original draft: B.S., B.M.C. and P.J.C. Writing, review and editing: all authors. Funding acquisition: F.S., G.J.B., B.M.C. and P.J.C. Supervision: F.S., G.J.B., B.M.C. and P.J.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
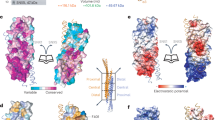
Supplementary Figure 1 Structures and electron density of the CI-MPR:SNX5 complex and comparison with the SNX5:IncE complex.
(A) For comparison the SNX5–IncE structure1 is shown in ribbon diagram (left panel), with a close-up of the bound IncE peptide in stick representation (right panel). (B) Comparison between the structures of SNX PX domain from the SNX5-IncE complex1, SNX32-IncE complex2 (PDB ID 6E8R2), SNX5–CI-MPR complex and the previously reported apo-SNX5 PX domain3 crystal structure (PDB ID 3HPB3).
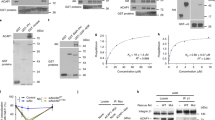
Supplementary Figure 2 Comparison between the interactome and the surface proteome of SNX5 and SNX5(F136D).
(A) Schematics of the TMT-labelling based proteomics and the approach used to filter and analyse the datasets. Raw proteomic data is provided in Supplementary Table 3. (B) Gene names of the proteins whose relative enrichment was statistically different in the SNX5(F136D) interactome compared to the SNX5 interactome. Analysis of comparative interactome of wild-type SNX5 vs SNX5(F136D) mutant across n = 3 independent experiments using One-sample t-test and Benjamini–Hochberg FDR. The unprocessed original proteomic datasets and the corresponding analysis are provided in Supplementary Table 3. (C) Schematics of surface biotinylation coupled to the TMT-labelling based proteomics and the approach used to filter and analyse the datasets. (D) Gene names of the proteins whose relative enrichment was statistically decreased in the SNX5(F136D) surface proteome compared to that of SNX5 wild-type. Analysis was performed across n = 3 independent experiments using One-sample t-test and Benjamini–Hochberg FDR. The unprocessed original proteomic datasets and the corresponding analysis are provided in Supplementary Table 3.
Supplementary Figure 3 Unprocessed original scans of immunoblots.
Unprocessed images of all gels and blots.
Supplementary information
Supplementary Information
Supplementary Figures 1–3, Supplementary Table titles/legends and Supplementary References.
Supplementary Table 1
Binding parameters from ITC experiments.
Supplementary Table 2
Summary of crystallographic structure determination statistics.
Supplementary Table 3
Unprocessed original proteomics and analysis.
Supplementary Table 4
Statistics source data.
Rights and permissions
About this article
Cite this article
Simonetti, B., Paul, B., Chaudhari, K. et al. Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins. Nat Cell Biol 21, 1219–1233 (2019). https://doi.org/10.1038/s41556-019-0393-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0393-3
This article is cited by
-
STAM and Hrs interact sequentially with IFN-α Receptor to control spatiotemporal JAK–STAT endosomal activation
Nature Cell Biology (2023)
-
Architecture of the ESCPE-1 membrane coat
Nature Structural & Molecular Biology (2023)
-
Phosphoinositides as membrane organizers
Nature Reviews Molecular Cell Biology (2022)
-
Recycling of autophagosomal components from autolysosomes by the recycler complex
Nature Cell Biology (2022)
-
FERARI and cargo adaptors coordinate cargo flow through sorting endosomes
Nature Communications (2022)