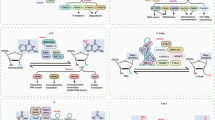
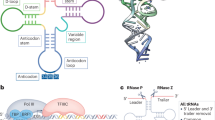
The deposition of chemical modifications into RNA is a crucial regulator of temporal and spatial gene expression programs during development. Accordingly, altered RNA modification patterns are widely linked to developmental diseases. Recently, the dysregulation of RNA modification pathways also emerged as a contributor to cancer. By modulating cell survival, differentiation, migration and drug resistance, RNA modifications add another regulatory layer of complexity to most aspects of tumourigenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291 (2017).
Frye, M., Jaffrey, S. R., Pan, T., Rechavi, G. & Suzuki, T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 17, 365–372 (2016).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Dominissini, D. et al. The dynamic N 1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Blanco, S. et al. Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340 (2016).
Natchiar, S. K., Myasnikov, A. G., Kratzat, H., Hazemann, I. & Klaholz, B. P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477 (2017).
Sloan, K. E. et al. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosomebiogenesis and function. RNA Biol. 14, 1138–1152 (2017).
Pan, T. Modifications and functional genomics of human transfer RNA. Cell Res. 28, 395–404 (2018).
Rich, A. & RajBhandary, U. L. Transfer RNA: molecular structure, sequence, and properties. Annu. Rev. Biochem. 45, 805–860 (1976).
Schimmel, P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 19, 45–58 (2018).
Adams, J. M. & Cory, S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature 255, 28–33 (1975).
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA 71, 3971–3975 (1974).
Dubin, D. T. & Taylor, R. H. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 2, 1653–1668 (1975).
Perry, R. P., Kelley, D. E., Friderici, K. & Rottman, F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell 4, 387–394 (1975).
Schibler, U., Kelley, D. E. & Perry, R. P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 115, 695–714 (1977).
Wei, C. M., Gershowitz, A. & Moss, B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4, 379–386 (1975).
Safra, M. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255 (2017).
Li, X. et al. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68, 993–1005.e1009 (2017).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Delatte, B. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Yang, X. et al. 5-Methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 27, 606–625 (2017).
Akichika, S. et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080 (2019).
Rintala-Dempsey, A. C. & Kothe, U. Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 14, 1185–1196 (2017).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Bokar, J. A., Rath-Shambaugh, M. E., Ludwiczak, R., Narayan, P. & Rottman, F. Characterization and partial purification of mRNA N 6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269, 17697–17704 (1994).
Ke, S. et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017).
Bertero, A. et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 555, 256–259 (2018).
Aguilo, F. et al. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 17, 689–704 (2015).
Barbieri, I. et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 552, 126–131 (2017).
Huang, H. et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567, 414–419 (2019).
Jia, G. et al. N 6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Wang, X. et al. N 6-Methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Ivanova, I. et al. The RNA m6A Reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell 67, 1059–1067.e1054 (2017).
Zhao, B. S. et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 (2017).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Shi, H. et al. YTHDF3 facilitates translation and decay of N 6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Wang, X. et al. N 6-Methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Bailey, A. S. et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6, e26116 (2017).
Huang, H. et al. Recognition of RNA N 6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Yang, Y., Hsu, P. J., Chen, Y.-S. & Yang, Y.-G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 28, 616–624 (2018).
Patil, D. P., Pickering, B. F. & Jaffrey, S. R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28, 113–127 (2018).
Roundtree, I. A. et al. YTHDC1 mediates nuclear export of N 6-methyladenosine methylated mRNAs. eLife 6, e31311 (2017).
Buszczak, M., Signer, R. A. & Morrison, S. J. Cellular differences in protein synthesis regulate tissue homeostasis. Cell 159, 242–251 (2014).
Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009).
Robichaud, N., Sonenberg, N., Ruggero, D. & Schneider, R.J. Translational control in cancer. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a032896 (2018).
Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016).
Dittmar, K. A., Goodenbour, J. M. & Pan, T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221 (2006).
Gingold, H. et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014).
Kuhn, C. D. RNA versatility governs tRNA function: why tRNA flexibility is essential beyond the translation cycle. BioEssays 38, 465–473 (2016).
Schaffrath, R. & Leidel, S. A. Wobble uridine modifications-a reason to live, a reason to die?! RNA Biol. 14, 1209–1222 (2017).
Hanson, G. & Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30 (2018).
Chan, C. T. et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3, 937 (2012).
Rezgui, V. A. N. et al. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl Acad. Sci. USA 110, 12289–12294 (2013).
Ranjan, N. & Rodnina, M. V. tRNA wobble modifications and protein homeostasis. Translation (Austin) 4, e1143076 (2016).
Rapino, F., Delaunay, S., Zhou, Z., Chariot, A. & Close, P. tRNA modification: is cancer having a wobble? Trends Cancer 3, 249–252 (2017).
Nedialkova, D. D. & Leidel, S. A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618 (2015).
Zinshteyn, B. & Gilbert, W. V. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 9, e1003675 (2013).
Blanco, S. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039 (2014).
Van Haute, L. et al. Deficient methylation and formylation of mt-tRNAMet wobble cytosine in a patientcarrying mutations in NSUN3. Nat. Commun. 7, 12039 (2016).
Nakano, S. et al. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet. Nat. Chem. Biol. 12, 546–551 (2016).
Haag, S. et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 35, 2104–2119 (2016).
Kawarada, L. et al. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 45, 7401–7415 (2017).
Thiaville, P. C. et al. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell 3, 29–45 (2016).
Tuorto, F. et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 34, 2350–2362 (2015).
Tuorto, F. et al. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 37, e99777 (2018).
Frye, M. & Blanco, S. Post-transcriptional modifications in development and stem cells. Development 143, 3871–3881 (2016).
Guzzi, N. et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216.e26 (2018).
Schaefer, M. et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24, 1590–1595 (2010).
Wang, X. et al. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313 (2018).
Torres, A. G., Batlle, E. & Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 20, 306–314 (2014).
Dai, D., Wang, H., Zhu, L., Jin, H. & Wang, X. N 6-Methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 9, 124 (2018).
Meacham, C. E. & Morrison, S. J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013).
Kreso, A. & Dick, J. E. Evolution of the cancer stem cell model. Cell Stem Cell 14, 275–291 (2014).
Li, Z. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N 6-methyladenosine RNA demethylase.Cancer Cell 31, 127–141 (2017).
Cui, Q. et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634 (2017).
Zhang, S. et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606.e596 (2017).
Vu, L. P. et al. The N 6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017).
Chen, M. et al. RNA N 6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67, 2254–2270 (2018).
Weng, H. et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 22, 191–205.e9 (2018).
Su, R. et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell 172, 90–105.e123 (2018).
Liu, J. et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 20, 1074–1083 (2018).
Rapino, F. et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 558, 605–609 (2018).
Delaunay, S. et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 213, 2503–2523 (2016).
Begley, U. et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α. EMBO Mol. Med. 5, 366–383 (2013).
Ladang, A. et al. Elp3 drives Wnt-dependent tumor initiation and regeneration in the intestine. J. Exp. Med. 212, 2057–2075 (2015).
Close, P. et al. DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator. J. Biol. Chem. 287, 32535–32545 (2012).
Merlos-Suárez, A. et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524 (2011).
Larsimont, J. C. et al. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell 17, 60–73 (2015).
Laguesse, S. et al. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell 35, 553–567 (2015).
Creppe, C. et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 136, 551–564 (2009).
Kojic, M. et al. Elongator mutation in mice induces neurodegeneration and ataxia-like behavior. Nat. Commun. 9, 3195 (2018).
Abbasi-Moheb, L. et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90, 847–855 (2012).
Khan, M. A. et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90, 856–863 (2012).
Martinez, F. J. et al. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet. 49, 380–385 (2012).
Frye, M. et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 289, 71–80 (2010).
Lu, L., Zhu, G., Zeng, H., Xu, Q. & Holzmann, K. High tRNA transferase NSUN2 gene expression is associated with poor prognosis in head and neck squamous carcinoma. Cancer Invest. 36, 246–253 (2018).
Signer, R. A., Magee, J. A., Salic, A. & Morrison, S. J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014).
Signer, R. A. et al. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev. 30, 1698–1703 (2016).
Llorens-Bobadilla, E. et al. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329–340 (2015).
Baser, A. et al. Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature 566, 100–104 (2019).
Wek, R. C., Jiang, H. Y. & Anthony, T. G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 (2006).
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Batista, P. J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Huang, W. et al. Determination of DNA and RNA methylation in circulating tumor cells by mass spectrometry. Anal. Chem. 88, 1378–1384 (2016).
Zhang, C. et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 113, E2047–E2056 (2016).
Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R. I. The m6A methyltransferase METTL3 promotestranslation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Deng, X. et al. RNA N 6-methyladenosine modification in cancers: current status and perspectives. Cell Res. 28, 507–517 (2018).
Popis, M. C., Blanco, S. & Frye, M. Posttranscriptional methylation of transfer and ribosomal RNA in stress response pathways, cell differentiation, and cancer. Curr. Opin. Oncol. 28, 65–71 (2016).
Dachs, G. U. & Tozer, G. M. Hypoxia modulated gene expression: angiogenesis, metastasis and therapeutic exploitation. Eur. J. Cancer 36, 1649–1660 (2000).
Rankin, E. B. & Giaccia, A. J. Hypoxic control of metastasis. Science 352, 175–180 (2016).
Chionh, Y. H. et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 7, 13302 (2016).
Flores, J. V. et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Reports 8, 112–124 (2017).
Yi, J. et al. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget 8, 20751–20765 (2017).
Goodarzi, H. et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161, 790–802 (2015).
Choe, J. et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560 (2018).
Ma, J. Z. et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N 6-methyladenosine-dependent primary MicroRNA processing. Hepatology 65, 529–543 (2017).
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N. & Tavazoie, S. F. N 6-Methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015).
Okamoto, M. et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 10, e1004639 (2014).
Alexandrov, A., Martzen, M. R. & Phizicky, E. M. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8, 1253–1266 (2002).
Longley, D. B., Harkin, D. P. & Johnston, P. G. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3, 330–338 (2003).
Blanco, S. et al. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 7, e1002403 (2011).
Ward, P. S. & Thompson, C. B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308 (2012).
Alexandrov, A. et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21, 87–96 (2006).
Rubio, M. A. et al. Editing and methylation at a single site by functionally interdependent activities. Nature 542, 494–497 (2017).
Phizicky, E. M. & Hopper, A. K. tRNA biology charges to the front. Genes Dev. 24, 1832–1860 (2010).
Björk, G. R. et al. Transfer RNA modification. Annu. Rev. Biochem. 56, 263–287 (1987).
Acknowledgements
M.F. is supported by a Cancer Research UK Senior Fellowship (C10701/A15181), the European Research Council (ERC; 310360), and the Medical Research Council UK (MR/M01939X/1). S.D. is funded by an EMBO Long-Term Fellowship. Part of this work was carried out in the framework of the European COST action EPITRAN 16120.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
M.F. consults for STORM Therapeutics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Delaunay, S., Frye, M. RNA modifications regulating cell fate in cancer. Nat Cell Biol 21, 552–559 (2019). https://doi.org/10.1038/s41556-019-0319-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0319-0
This article is cited by
-
Age-related noncanonical TRMT6–TRMT61A signaling impairs hematopoietic stem cells
Nature Aging (2024)
-
Emerging role of RNA modification and long noncoding RNA interaction in cancer
Cancer Gene Therapy (2024)
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)
-
YTHDF2 facilitates aggresome formation via UPF1 in an m6A-independent manner
Nature Communications (2023)
-
Regulation and functions of non-m6A mRNA modifications
Nature Reviews Molecular Cell Biology (2023)