Abstract
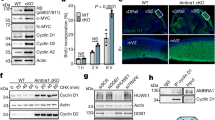
Growth signals, such as extracellular nutrients and growth factors, have substantial effects on genome integrity; however, the direct underlying link remains unclear. Here, we show that the mechanistic target of rapamycin (mTOR)–ribosomal S6 kinase (S6K) pathway, a central regulator of growth signalling, phosphorylates RNF168 at Ser60 to inhibit its E3 ligase activity, accelerate its proteolysis and impair its function in the DNA damage response, leading to accumulated unrepaired DNA and genome instability. Moreover, loss of the tumour suppressor liver kinase B1 (LKB1; also known as STK11) hyperactivates mTOR complex 1 (mTORC1)–S6K signalling and decreases RNF168 expression, resulting in defects in the DNA damage response. Expression of a phospho-deficient RNF168-S60A mutant rescues the DNA damage repair defects and suppresses tumorigenesis caused by Lkb1 loss. These results reveal an important function of mTORC1–S6K signalling in the DNA damage response and suggest a general mechanism that connects cell growth signalling to genome stability control.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Langie, S. A. et al. Causes of genome instability: the effect of low dose chemical exposures in modern society. Carcinogenesis 36, S61–S88 (2015).
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell. 40, 179–204 (2010).
Symington, L. S. & Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 (2011).
Khanna, K. K. & Jackson, S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 (2001).
Jackson, S. P. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23, 687–696 (2002).
Huen, M. S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007).
Kolas, N. K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007).
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009).
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Heydari, A. R., Unnikrishnan, A., Lucente, L. V. & Richardson, A. Caloric restriction and genomic stability. Nucleic Acids Res. 35, 7485–7496 (2007).
Szafranski, K. & Mekhail, K. The fine line between lifespan extension and shortening in response to caloric restriction. Nucleus 5, 56–65 (2014).
Esteve-Puig, R. et al. A mouse model uncovers LKB1 as an UVB-induced DNA damage sensor mediating CDKN1A (p21WAF1/CIP1) degradation. PLoS. Genet. 10, e1004721 (2014).
Ui, A. et al. Possible involvement of LKB1–AMPK signaling in non-homologous end joining. Oncogene 33, 1640–1648 (2014).
Gupta, R., Liu, A. Y., Glazer, P. M. & Wajapeyee, N. LKB1 preserves genome integrity by stimulating BRCA1 expression. Nucleic Acids Res. 43, 259–271 (2015).
Alexander, A. et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl Acad. Sci. USA 107, 4153–4158 (2010).
Tripathi, D. N. et al. Reactive nitrogen species regulate autophagy through ATM–AMPK–TSC2-mediated suppression of mTORC1. Proc. Natl Acad. Sci. USA 110, E2950–E2957 (2013).
Shen, C. et al. Regulation of FANCD2 by the mTOR pathway contributes to the resistance of cancer cells to DNA double-strand breaks. Cancer Res. 73, 3393–3401 (2013).
Johnson, S. C., Rabinovitch, P. S. & Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 493, 338–345 (2013).
Shen, C. et al. TOR signaling is a determinant of cell survival in response to DNA damage. Mol. Cell. Biol. 27, 7007–7017 (2007).
Kim, D. H. et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002).
Shimobayashi, M. & Hall, M. N. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell. Biol. 15, 155–162 (2014).
Weinstock, D. M., Nakanishi, K., Helgadottir, H. R. & Jasin, M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 409, 524–540 (2006).
Korsse, S. E., Peppelenbosch, M. P. & van Veelen, W. Targeting LKB1 signaling in cancer. Biochim. Biophys. Acta 1835, 194–210 (2013).
Al-Hakim, A. et al. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair 9, 1229–1240 (2010).
Mattiroli, F. et al. RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012).
Ferrari, S., Bandi, H. R., Hofsteenge, J., Bussian, B. M. & Thomas, G. Mitogen-activated 70K S6 kinase. Identification of in vitro 40 S ribosomal S6 phosphorylation sites. J. Biol. Chem. 266, 22770–22775 (1991).
Gudjonsson, T. et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150, 697–709 (2012).
Shackelford, D. B. & Shaw, R. J. The LKB1–AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575 (2009).
Alexander, A. & Walker, C. L. The role of LKB1 and AMPK in cellular responses to stress and damage. FEBS Lett. 585, 952–957 (2011).
De Raedt, T. et al. Exploiting cancer cell vulnerabilities to develop a combination therapy for Ras-driven tumors. Cancer Cell. 20, 400–413 (2011).
Calles, A. et al. Immunohistochemical loss of LKB1 is a biomarker for more aggressive biology in KRAS-mutant lung adenocarcinoma. Clin. Cancer Res. 21, 2851–2860 (2015).
Ji, H. et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 448, 807–810 (2007).
Bandhakavi, S. et al. Quantitative nuclear proteomics identifies mTOR regulation of DNA damage response. Mol. Cell. Proteom. 9, 403–414 (2010).
Schieber, M. & Chandel, N. S. ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462 (2014).
Damsky, W. et al. mTORC1 activation blocks Braf V600E-induced growth arrest but is insufficient for melanoma formation. Cancer Cell. 27, 41–56 (2015).
Cantor, J. R. & Sabatini, D. M. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2, 881–898 (2012).
Martinez-Outschoorn, U. E., Peiris-Pages, M., Pestell, R. G., Sotgia, F. & Lisanti, M. P. Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 14, 11–31 (2017).
Wellen, K. E. & Thompson, C. B. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol. Cell. 40, 323–332 (2010).
Katajisto, P. et al. The LKB1 tumor suppressor kinase in human disease. Biochim. Biophys. Acta 1775, 63–75 (2007).
Inoki, K. et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–968 (2006).
Gwinn, D. M. et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 30, 214–226 (2008).
van Veelen, W., Korsse, S. E., van de Laar, L. & Peppelenbosch, M. P. The long and winding road to rational treatment of cancer associated with LKB1/AMPK/TSC/mTORC1 signaling. Oncogene 30, 2289–2303 (2011).
Liu, P. et al. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat. Cell. Biol. 15, 1340–1350 (2013).
Boehm, J. S., Hession, M. T., Bulmer, S. E. & Hahn, W. C. Transformation of human and murine fibroblasts without viral oncoproteins. Mol. Cell. Biol. 25, 6464–6474 (2005).
Gao, D. et al. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat. Cell. Biol. 11, 397–408 (2009).
Gao, D. et al. Cdh1 regulates cell cycle through modulating the claspin/Chk1 and the Rb/E2F1 pathways. Mol. Biol. Cell. 20, 3305–3316 (2009).
Franken, N. A., Rodermond, H. M., Stap, J., Haveman, J. & van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 1, 2315–2319 (2006).
Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes. Dev. 13, 2633–2638 (1999).
Tanaka, Y. et al. Expression and purification of recombinant human histones. Methods 33, 3–11 (2004).
Olive, P. L. & Banath, J. P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 1, 23–29 (2006).
Liu, C. et al. RNF168 forms a functional complex with RAD6 during the DNA damage response. J. Cell. Sci. 126, 2042–2051 (2013).
Bohgaki, T. et al. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS. Genet. 7, e1001381 (2011).
Acknowledgements
We thank D. Durocher, C. Lucas and L. Penengo for sharing RNF168 constructs, and K.-L. Guan and D. Li for critical reading of the manuscript. This work was supported by the National Key Basic Research Program of China (2015CB964502), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), and grants from the National Science foundation of China to D.G. (81372602 and 81422033) and to Xiaoduo Xie (31401214). P.L. is supported by 1K99CA181342 and 5T32HL007893 from National Institutes of Health (USA).
Author information
Authors and Affiliations
Contributions
Xiaoduo Xie, H.H., X.T. and L.L. performed most of the experiments with assistance from X.L., M.C., Xia Xie, Y.Z., H.O., M.W., J.H., P.L., W.G., Y.L., A.X., X.K. and G.-W.C. H.Y. performed the villi length assay of mice after IR treatment under the supervision of J.Q. Q.L. and H.Z. performed the mass spectrometry analysis under the supervision of R.Z. D.G., Xiaoduo Xie, H.H.,W.W. and H.J. designed the experiments with input from R.H. F.-L.M., D.G., Xiaoduo Xie and H.H. wrote the manuscript. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–8, Supplementary Table legends.
Supplementary Table 1
Primary antibodies and reagents used in this study.
Supplementary Table 2
Target sequences of shRNAs and siRNAs used in this study.
Supplementary Table 3
Statistics source data.
Rights and permissions
About this article
Cite this article
Xie, X., Hu, H., Tong, X. et al. The mTOR–S6K pathway links growth signalling to DNA damage response by targeting RNF168. Nat Cell Biol 20, 320–331 (2018). https://doi.org/10.1038/s41556-017-0033-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-017-0033-8
This article is cited by
-
Selenoproteins synergistically protect porcine skeletal muscle from oxidative damage via relieving mitochondrial dysfunction and endoplasmic reticulum stress
Journal of Animal Science and Biotechnology (2023)
-
Expression of mTOR in normal and pathological conditions
Molecular Cancer (2023)
-
Beyond controlling cell size: functional analyses of S6K in tumorigenesis
Cell Death & Disease (2022)
-
AMPK induces degradation of the transcriptional repressor PROX1 impairing branched amino acid metabolism and tumourigenesis
Nature Communications (2022)
-
Aspirin sensitivity of PIK3CA-mutated Colorectal Cancer: potential mechanisms revisited
Cellular and Molecular Life Sciences (2022)