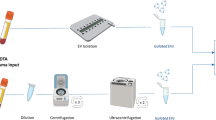
Abstract
Assays for cancer diagnosis via the analysis of biomarkers on circulating extracellular vesicles (EVs) typically have lengthy sample workups, limited throughput or insufficient sensitivity, or do not use clinically validated biomarkers. Here we report the development and performance of a 96-well assay that integrates the enrichment of EVs by antibody-coated magnetic beads and the electrochemical detection, in less than one hour of total assay time, of EV-bound proteins after enzymatic amplification. By using the assay with a combination of antibodies for clinically relevant tumour biomarkers (EGFR, EpCAM, CD24 and GPA33) of colorectal cancer (CRC), we classified plasma samples from 102 patients with CRC and 40 non-CRC controls with accuracies of more than 96%, prospectively assessed a cohort of 90 patients, for whom the burden of tumour EVs was predictive of five-year disease-free survival, and longitudinally analysed plasma from 11 patients, for whom the EV burden declined after surgery and increased on relapse. Rapid assays for the detection of combinations of tumour biomarkers in plasma EVs may aid cancer detection and patient monitoring.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw patient datasets generated and analysed during the study are available from the corresponding authors on reasonable request, subject to approval from the Institutional Review Board of the Kyungpook National University Hospital. For initial marker selection, we used the public databases, the Human Protein Atlas (https://www.proteinatlas.org) and UniProt (https://www.uniprot.org). Source data are provided with this paper.
Code availability
Source codes for the marker selection are available at https://csb.mgh.harvard.edu/bme_software.
References
Heitzer, E., Haque, I. S., Roberts, C. E. S. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 77–88 (2018).
Siravegna, G., Marsoni, S., Siena, S. & Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548 (2017).
Chi, K. R. The tumour trail left in blood. Nature 532, 269–271 (2016).
Pantel, K. & Alix-Panabieres, C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 73, 6384–6388 (2013).
Théry, C., Ostrowski, M. & Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 (2009).
Xu, R. et al. Extracellular vesicles in cancer—implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 15, 617–638 (2018).
Shao, H. et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 18, 1835–1840 (2012).
Choi, D., Spinelli, C., Montermini, L. & Rak, J. Oncogenic regulation of extracellular vesicle proteome and heterogeneity. Proteomics 19, 1800169 (2019).
Skog, J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 (2008).
Shao, H. et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 6, 6999 (2015).
Yoshioka, Y. et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun. 5, 3591 (2014).
Skotland, T., Sandvig, K. & Llorente, A. Lipids in exosomes: current knowledge and the way forward. Prog. Lipid Res. 66, 30–41 (2017).
EL Andaloussi, S., Mäger, I., Breakefield, X. O. & Wood, M. J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357 (2013).
Im, H. et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 32, 490–495 (2014).
Liu, C. et al. Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers. Nat. Biomed. Eng. 3, 183–193 (2019).
Jeong, S. et al. Integrated magneto–electrochemical sensor for exosome analysis. ACS Nano 10, 1802–1809 (2016).
Yang, K. S. et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci. Transl. Med. 9, eaal3226 (2017).
Zhang, P. et al. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 3, 438–451 (2019).
Liang, K. et al. Nanoplasmonic quantification of tumour-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat. Biomed. Eng. 1, 0021 (2017).
Lewis, J. M. et al. Integrated analysis of exosomal protein biomarkers on alternating current electrokinetic chips enables rapid detection of pancreatic cancer in patient blood. ACS Nano 12, 3311–3320 (2018).
Shao, H. et al. New technologies for analysis of extracellular vesicles. Chem. Rev. 118, 1917–1950 (2018).
Locker, G. Y. et al. ASCO 2006 update of recommendations for the use of tumour markers in gastrointestinal cancer. J. Clin. Oncol. 24, 5313–5327 (2006).
Tao, S., Hundt, S., Haug, U. & Brenner, H. Sensitivity estimates of blood-based tests for colorectal cancer detection: impact of overrepresentation of advanced stage disease. Am. J. Gastroenterol. 106, 242–253 (2011).
Park, J. et al. Integrated kidney exosome analysis for the detection of kidney transplant rejection. ACS Nano 11, 11041–11046 (2017).
Fraser, K. et al. Characterization of single microvesicles in plasma from glioblastoma patients. Neuro Oncol. 21, 606–615 (2019).
Jeppesen, D. K. et al. Reassessment of exosome composition. Cell 177, 428–445 (2019).
Lee, K. et al. Multiplexed profiling of single extracellular vesicles. ACS Nano 12, 494–503 (2018).
Ramirez, M. I. et al. Technical challenges of working with extracellular vesicles. Nanoscale 10, 881–906 (2018).
Zhao, L. H. et al. CD44v6 expression in patients with stage II or stage III sporadic colorectal cancer is superior to CD44 expression for predicting progression. Int. J. Clin. Exp. Pathol. 8, 692–701 (2015).
Weichert, W. et al. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin. Cancer Res. 11, 6574–6581 (2005).
Weichert, W., Knösel, T., Bellach, J., Dietel, M. & Kristiansen, G. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J. Clin. Pathol. 57, 1160–1164 (2004).
Lee, C. H. et al. The prognostic role of STEAP1 expression determined via immunohistochemistry staining in predicting prognosis of primary colorectal cancer: a survival analysis. Int. J. Mol. Sci. 17, 592 (2016).
Ingebrigtsen, V. A. et al. B7-H3 expression in colorectal cancer: nuclear localization strongly predicts poor outcome in colon cancer. Int. J. Cancer 131, 2528–2536 (2012).
Deng, Y. et al. ALDH1 is an independent prognostic factor for patients with stages II-III rectal cancer after receiving radiochemotherapy. Br. J. Cancer 110, 430–434 (2014).
Ong, C. W. et al. CD133 expression predicts for non-response to chemotherapy in colorectal cancer. Mod. Pathol. 23, 450–457 (2010).
Peters, G. J. et al. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim. Biophys. Acta 1587, 194–205 (2002).
Ekblad, L., Kjellström, J. & Johnsson, A. Reduced drug accumulation is more important in acquired resistance against oxaliplatin than against cisplatin in isogenic colon cancer cells. Anticancer Drugs 21, 523–531 (2010).
Mouradov, D. et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 74, 3238–3247 (2014).
Skog, J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470 (2008).
Garinchesa, P. et al. Organ-specific expression of the colon cancer antigen A33, a cell surface target for antibody-based therapy. Int. J. Oncol. 9, 465–471 (1996).
Zou, K. H., O’Malley, A. J. & Mauri, L. Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation 115, 654–657 (2007).
Liu, J. et al. Down-regulation of GADD45A enhances chemosensitivity in melanoma. Sci. Rep. 8, 4111 (2018).
Huang, P. et al. Chemotherapy-driven increases in the CDKN1A/PTN/PTPRZ1 axis promote chemoresistance by activating the NF-κB pathway in breast cancer cells. Cell Commun. Signal. 16, 92 (2018).
Cuadrado, A. et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 18, 295–317 (2019).
Derdak, Z. et al. The mitochondrial uncoupling protein-2 promotes chemoresistance in cancer cells. Cancer Res. 68, 2813–2819 (2008).
Nicolussi, A., D’Inzeo, S., Capalbo, C., Giannini, G. & Coppa, A. The role of peroxiredoxins in cancer. Mol. Clin. Oncol. 6, 139–153 (2017).
Wang, J. & Li, Y. CD36 tango in cancer: signaling pathways and functions. Theranostics 9, 4893–4908 (2019).
Romano, G. et al. The TGF-β pathway is activated by 5-fluorouracil treatment in drug resistant colorectal carcinoma cells. Oncotarget 7, 22077–22091 (2016).
Wu, J. et al. Heat shock proteins and cancer. Trends Pharmacol. Sci. 38, 226–256 (2017).
Sharma, A., Upadhyay, A. K. & Bhat, M. K. Inhibition of Hsp27 and Hsp40 potentiates 5-fluorouracil and carboplatin mediated cell killing in hepatoma cells. Cancer Biol. Ther. 8, 2106–2113 (2009).
Shi, Z. et al. Activation of the PERK-ATF4 pathway promotes chemo-resistance in colon cancer cells. Sci. Rep. 9, 3210 (2019).
Phallen, J. et al. Direct detection of early-stage cancers using circulating tumour DNA. Sci. Transl. Med. 9, eaan2415 (2017).
Hothorn, T. & Lausen, B. On the exact distribution of maximally selected rank statistics. Comput. Stat. Data Anal. 43, 121–137 (2003).
Duffy, M. J. et al. Clinical utility of biochemical markers in colorectal cancer. Eur. J. Cancer 39, 718–727 (2003).
Das, J. et al. An electrochemical clamp assay for direct, rapid analysis of circulating nucleic acids in serum. Nat. Chem. 7, 569–575 (2015).
Thierry, A. R. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumour DNA. Nat. Med. 20, 430–435 (2014).
Network, C. G. A. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Siravegna, G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 21, 795–801 (2015).
Hoshino, A. et al. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015).
Peinado, H. et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017).
Keklikoglou, I. et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat. Cell Biol. 21, 190–202 (2019).
Syn, N., Wang, L., Sethi, G., Thiery, J. P. & Goh, B. C. Exosome-mediated metastasis: from epithelial-mesenchymal transition to escape fromimmunosurveillance. Trends Pharmacol. Sci. 37, 606–617 (2016).
Van Deun, J. et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 3, 24858 (2014).
DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845 (1988).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011).
Hothorn, T. & Lausen, B. On the exact distribution of maximally selected rank statistics. Comp. Stat. Data Analysis 43, 121–137 (2003).
Acknowledgements
We thank X. O. Breakefield (Massachusetts General Hospital) for discussions. This work was supported in part by grants from the V-Foundation for Cancer Research (R. Weissleder, C.M.C.); the American Cancer Society (C.M.C.); the Robert Wood Johnson Foundation/Amos Medical Faculty Development Program (C.M.C.); US National Institutes of Health (NIH) grant numbers R01CA229777 (H. Lee), R21DA049577 (H. Lee), R01CA204019 (R. Weissleder), U01CA233360 (H. Lee, C.M.C.), TR000931 (B.S.C.), U01CA230697 (B.S.C., L.B.), R01CA239078 (H. Lee, B.S.C.), R01CA237500 (H. Lee, B.S.C.) and CA069246 (B.S.C.); US DOD-W81XWH1910199 (H. Lee) and DOD-W81XWH1910194 (H. Lee); MGH Scholar Fund (H. Lee), MGH Fund for Medical Discovery Fellowship (H.-Y.L.); and Basic Science Research Program grants NRF-2019R1C1C1008792 (J.P.), NRF-2020R1A4A1016093 (J.P.), and NRF-2017M3A9G8083382 (J.S.P.) from the Ministry of Education, South Korea.
Author information
Authors and Affiliations
Contributions
J.P., J.S.P., C.M.C., R. Weissleder and H. Lee designed the study, prepared the figures and wrote the manuscript. J.S.P. and G.-S.C. collected patient samples. G.Y. performed tissue immunohistochemistry staining. C.-H.H. developed the HiMEX system. J.P., A.J., H.-Y.L., J.V.D., H. Li and J.M. performed research and analysed data. L.W., L.B. and B.S.C. guided mRNA analysis. J.S.P., G.-S.C., C.M.C. and R. Weissleder analysed clinical data. K.C., R. Wang and H. Lee performed statistical analyses.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the filing of a patent (US20190346434A1) that was assigned to and handled by Massachusetts General Hospital. The following disclosures are not related to the subject matter of this work. R.Weissleder is a consultant to ModeRNA, Tarveda Pharmaceuticals, Lumicell, Seer, Earli, Alivio Therapeutics, Aikili Biosystems and Accure Health. H. Lee is a consultant to Exosome Diagnostics, Accure Health and Aikili Biosystems.
Additional information
Peer review information Nature Biomedical Engineering thanks Tony Hu, Philip Stahl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures and tables.
Source data
Source data for Fig. 2
Data for Fig. 2c–e.
Rights and permissions
About this article
Cite this article
Park, J., Park, J.S., Huang, CH. et al. An integrated magneto-electrochemical device for the rapid profiling of tumour extracellular vesicles from blood plasma. Nat Biomed Eng 5, 678–689 (2021). https://doi.org/10.1038/s41551-021-00752-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00752-7
This article is cited by
-
Extracellular vesicle-based liquid biopsy biomarkers and their application in precision immuno-oncology
Biomarker Research (2023)
-
Ratiometric electrochemical OR gate assay for NSCLC-derived exosomes
Journal of Nanobiotechnology (2023)
-
Advanced extracellular vesicle bioinformatic nanomaterials: from enrichment, decoding to clinical diagnostics
Journal of Nanobiotechnology (2023)
-
Analytical device miniaturization for the detection of circulating biomarkers
Nature Reviews Bioengineering (2023)
-
Comprehensive isolation of extracellular vesicles and nanoparticles
Nature Protocols (2023)