Abstract
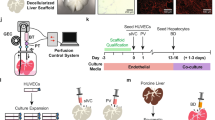
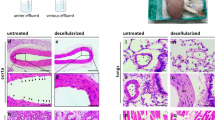
Implanted bioengineered livers have not exceeded three days of continuous perfusion. Here we show that decellularized whole porcine livers revascularized with human umbilical vein endothelial cells and implanted heterotopically into immunosuppressed pigs whose spleens had been removed can sustain perfusion for up to 15 days. We identified peak glucose consumption rate as a main predictor of the patency of the revascularized bioengineered livers (rBELs). Heterotopic implantation of rBELs into pigs in the absence of anticoagulation therapy led to sustained perfusion for three days, followed by a pronounced immune responses directed against the human endothelial cells. A 10 day steroid-based immunosuppression protocol and a splenectomy at the time of rBEL implantation reduced the immune responses and resulted in continuous perfusion of the rBELs for over two weeks. We also show that the human endothelial cells in the perfused rBELs colonize the liver sinusoids and express sinusoidal endothelial markers similar to those in normal liver tissue. Revascularized liver scaffolds that can maintain blood perfusion at physiological pressures might eventually help to overcome the chronic shortage of transplantable human livers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The main data supporting the results in this study are available in the Article and Supplementary Information. The raw and analysed datasets generated during the study are available for research purposes from the corresponding authors on reasonable request.
Change history
05 November 2019
A Correction to this paper has been published: https://doi.org/10.1038/s41551-019-0483-3
References
Shirakigawa, N., Takei, T. & Ijima, H. Base structure consisting of an endothelialized vascular-tree network and hepatocytes for whole liver engineering. J. Biosci. Bioeng. 116, 740–745 (2013).
Badylak, S. F., Taylor, D. & Uygun, K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu. Rev. Biomed. Eng. 13, 27–53 (2011).
Soto-Gutierrez, A. et al. A whole-organ regenerative medicine approach for liver replacement. Tissue Eng. Pt C 17, 677–686 (2011).
Baptista, P. M. et al. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology 53, 604–617 (2011).
Barakat, O. et al. Use of decellularized porcine liver for engineering humanized liver organ. J. Surg. Res. 173, e11–e25 (2012).
Bao, J. et al. Construction of a portal implantable functional tissue-engineered liver using perfusion-decellularized matrix and hepatocytes in rats. Cell Transplant. 20, 753–766 (2011).
Zhou, P. et al. Decellularized liver matrix as a carrier for the transplantation of human fetal and primary hepatocytes in mice. Liver Transplant. 17, 418–427 (2011).
Yagi, H. et al. Human-scale whole-organ bioengineering for liver transplantation: a regenerative medicine approach. Cell Transplant. 22, 231–242 (2013).
Uygun, B. E. et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat. Med. 16, 814–820 (2010).
Bao, J. et al. Hemocompatibility improvement of perfusion-decellularized clinical-scale liver scaffold through heparin immobilization. Sci. Rep. 5, 10756 (2015).
Park, K. M. et al. Decellularized liver extracellular matrix as promising tools for transplantable bioengineered liver promotes hepatic lineage commitments of induced pluripotent stem cells. Tissue Eng. Pt A 22, 449–460 (2016).
Wang, Y. et al. Recent advances in decellularization and recellularization for tissue-engineered liver grafts. Cells Tissues Organs 204, 125–136 (2017).
Uygun, B. E. & Yarmush, M. L. Engineered liver for transplantation. Curr. Opin. Biotechnol. 24, 893–899 (2013).
Mirmalek-Sani, S.-H., Sullivan, D. C., Zimmerman, C., Shupe, T. D. & Petersen, B. E. Immunogenicity of decellularized porcine liver for bioengineered hepatic tissue. Am. J. Pathol. 183, 558–565 (2013).
Uzarski, J. S. et al. Dual-purpose bioreactors to monitor noninvasive physical and biochemical markers of kidney and liver scaffold recellularization. Tissue Eng. Pt C 21, 1032–1043 (2015).
Robertson, M. J., Soibam, B., O’Leary, J. G., Sampaio, L. C. & Taylor, D. A. Recellularization of rat liver: an in vitro model for assessing human drug metabolism and liver biology. PLoS ONE 13, e0191892 (2018).
Mazza, G., Al-Akkad, W., Rombouts, K. & Pinzani, M. Liver tissue engineering: from implantable tissue to whole organ engineering. Hepatol. Commun. 2, 131–141 (2018).
Rogers, S. C., Zhang, X., Azhar, G., Luo, S. & Wei, J. Y. Exposure to high or low glucose levels accelerates the appearance of markers of endothelial cell senescence and induces dysregulation of nitric oxide synthase. J. Gerontol. A 68, 1469–1481 (2013).
Lalor, P. F., Lai, W. K., Curbishley, S. M., Shetty, S. & Adams, D. H. Human hepatic sinusoidal endothelial cells can be distinguished by expression of phenotypic markers related to their specialised functions in vivo. World J. Gastroenterol. 12, 5429–5439 (2006).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
DeLeve, L. D. Liver sinusoidal endothelial cells in hepatic fibrosis. Hepatology 61, 1740–1746 (2015).
Skovseth, D. K., Yamanaka, T., Brandtzaeg, P., Butcher, E. C. & Haraldsen, G. Vascular morphogenesis and differentiation after adoptive transfer of human endothelial cells to immunodeficient mice. Am. J. Pathol. 160, 1629–1637 (2002).
Leach, L., Hamilton, R. D. & Foss, A. J. E. Phenotypic plasticity of human umbilical vein endothelial cells. Br. J. Ophthalmol. 96, 1152 (2012).
Robb, R. A. The biomedical imaging resource at Mayo Clinic. IEEE Trans. Med. Imaging 20, 854–867 (2001).
Nyberg, S. L., Amiot, B., Hardin, J., Baskin-Bey, E. & Platt, J. L. Cytotoxic immune response to a xenogeneic bioartificial liver. Cell Transplant. 13, 783–792 (2004).
Kin, W. R. et al. OPTN/SRTR 2016 annual data report: liver. Am. J. Transplant. 18, 172–253 (2018).
Lopez, P. M. & Martin, P. Update on liver transplantation: indications, organ allocation, and long-term care. Mt Sinai J. Med. 73, 1056–1066 (2006).
Dhawan, A., Puppi, J., Hughes, R. D. & Mitry, R. R. Human hepatocyte transplantation: current experience and future challenges. Nat. Rev. Gastroenterol. Hepatol. 7, 288–298 (2010).
Suchy, F., Yamaguchi, T. & Nakauchi, H. iPSC-derived organs in vivo: challenges and promise. Cell Stem Cell 22, 21–24 (2018).
Nicolas, C., Wang, Y. & Nyberg, S. L. Cell therapy in chronic liver disease. Curr. Opin. Gastroenterol. 32, 189–194 (2016).
Hussein, K. H., Park, K. M., Kang, K. S. & Woo, H. M. Heparin-gelatin mixture improves vascular reconstruction efficiency and hepatic function in bioengineered livers. Acta Biomater. 38, 82–93 (2016).
Ko, I. K. et al. Bioengineered transplantable porcine livers with re-endothelialized vasculature. Biomaterials 40, 72–79 (2015).
Mei, J. et al. The angiogenesis in decellularized scaffold-mediated the renal regeneration. Oncotarget 7, 27085–27093 (2016).
Guan, Y. et al. The effective bioengineering method of implantation decellularized renal extracellular matrix scaffolds. Oncotarget 6, 36126–36138 (2015).
Doi, R. et al. Transplantation of bioengineered rat lungs recellularized with endothelial and adipose-derived stromal cells. Sci. Rep. 7, 8447 (2017).
Mao, S., Glorioso, J., Elgilani, F., De Lorenzo, S. & Deeds, M. Sustained in vivo perfusion of a re-endothelialized tissue engineered porcine liver. Int. J. Transplant. Res. Med. 3, 031 (2017).
Dudakovic, A. et al. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J. Cell. Biochem. 115, 1816–1828 (2014).
Kalari, K. R. et al. MAP-RSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinformatics 15, 224 (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Metsalu, T. & Vilo, J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43, W566–W570 (2015).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
We thank C. Verfaillie and M. Kumar from the Stem Cell Institute Leuven, KU Leuven, Belgium for the qPCR analysis performed in the manuscript, the Mayo Clinic Microscopy and Cell Analysis Core for experimental and technical support and the Mayo Clinic Biomedical Imaging Resource for creating the 3D visualizations from CT images. We also thank L. Wentz for assistance with cell culture and J. Uzarski and M. M. Macenski for their review of and comments on the final manuscript. This work was made possible by financial support from Miromatrix, Mayo Clinic ILP grants and the National Institutes of Health (grant no. R01DK106667 to S.L.N.; grant nos. R01DK117861 and R03DK113339 to R.C.H.).
Author information
Authors and Affiliations
Contributions
M.F.S., J.J.R., D.S.D. and S.L.N. designed the study. A.Y. and V.Z. performed the HUVEC and rBEL culture. M.F.S., D.J.J., H.S.C., Y.L., B.A., E.N., T.M. and S.L.N. performed the surgical procedures. G.M. performed the CT. B.A., R.C.H. and M.L. performed the electron microscopy, B.G.S. performed the light and fluorescence microscopy. A.J.v.W. and C.R.P. performed the RNA-seq analysis. M.F.S., D.J.J., J.J.R., B.D.A., D.S.D., R.C.H., V.H.S., M.L. and S.L.N. analysed the experimental data. B.D.A. drafted the figures. M.F.S., J.J.R., B.D.A. and S.L.N. wrote the manuscript. R.C.H., V.H.S., A.B.D. and M.L. reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Miromatrix Medical Inc. is a privately funded company and owns the patent rights for the perfusion decellularization and recellularization technologies employed in this study. The research was funded by Miromatrix and a Mayo Clinic Innovation grant. J.J.R., B.D.A., A.Y., B.G.S. and D.S.D. are employees of Miromatrix.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Information
Supplementary figures and tables.
Supplementary Dataset 1
RNA-seq DAVID analysis.
Supplementary Dataset 2
Upregulated genes.
Supplementary Dataset 3
Similarity matrix input genes.
Supplementary Video 1
Intraoperative footage of the perfusion of the BEL graft.
Supplementary Video 2
3D CT reconstruction animation depicting the postoperative anatomy of the rBEL.
Rights and permissions
About this article
Cite this article
Shaheen, M.F., Joo, D.J., Ross, J.J. et al. Sustained perfusion of revascularized bioengineered livers heterotopically transplanted into immunosuppressed pigs. Nat Biomed Eng 4, 437–445 (2020). https://doi.org/10.1038/s41551-019-0460-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-019-0460-x
This article is cited by
-
Bioengineered liver crosslinked with nano-graphene oxide enables efficient liver regeneration via MMP suppression and immunomodulation
Nature Communications (2023)
-
Surface modification of decellularized bovine carotid arteries with human vascular cells significantly reduces their thrombogenicity
Journal of Biological Engineering (2021)
-
Generation of vascular chimerism within donor organs
Scientific Reports (2021)
-
Functional characterization of a bioengineered liver after heterotopic implantation in pigs
Communications Biology (2021)