Abstract
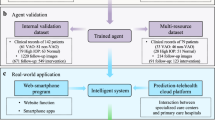
Using artificial intelligence (AI) to prevent and treat diseases is an ultimate goal in computational medicine. Although AI has been developed for screening and assisted decision-making in disease prevention and management, it has not yet been validated for systematic application in the clinic. In the context of rare diseases, the main strategy has been to build specialized care centres; however, these centres are scattered and their coverage is insufficient, which leaves a large proportion of rare-disease patients with inadequate care. Here, we show that an AI agent using deep learning, and involving convolutional neural networks for diagnostics, risk stratification and treatment suggestions, accurately diagnoses and provides treatment decisions for congenital cataracts in an in silico test, in a website-based study, in a ‘finding a needle in a haystack’ test and in a multihospital clinical trial. We also show that the AI agent and individual ophthalmologists perform equally well. Moreover, we have integrated the AI agent with a cloud-based platform for multihospital collaboration, designed to improve disease management for the benefit of patients with rare diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Patel, V. L. et al. The coming of age of artificial intelligence in medicine. Artif. Intel. Med. 46, 5–17 (2009).
Laksanasopin, T. et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 7, 273re1 (2015).
Kelly-Hope, L. A., Cano, J., Stanton, M. C., Bockarie, M. J. & Molyneux, D. H. Innovative tools for assessing risks for severe adverse events in areas of overlapping Loa loa and other filarial distributions: the application of micro-stratification mapping. Parasit. Vect. 7, 307 (2014).
Castaneda, C. et al. Clinical decision support systems for improving diagnostic accuracy and achieving precision medicine. J. Clin. Bioinf. 5, 4 (2015).
Schieppati, A., Henter, J. I., Daina, E. & Aperia, A. Why rare diseases are an important medical and social issue. Lancet 371, 2039–2041 (2008).
Taruscio, D., Vittozzi, L. & Stefanov, R. National plans and strategies on rare diseases in Europe. Adv. Exp. Med. Biol. 686, 475–491 (2010).
Remuzzi, G. & Garattini, S. Rare diseases: what's next? Lancet 371, 1978–1979 (2008).
Zhang, Y. J., Wang, Y. O., Li, L., Guo, J. J. & Wang, J. B. China’s first rare-disease registry is under development. Lancet 378, 769–770 (2011).
Gong, S. & Jin, S. Current progress in the management of rare diseases and orphan drugs in China. Intractable Rare Dis. Res. 1, 45–52 (2012).
Medsinge, A. & Nischal, K. K. Pediatric cataract: challenges and future directions. Clin. Ophthal. 9, 77–90 (2015).
Lin, H. et al. Lens regeneration using endogenous stem cells with gain of visual function. Nature 531, 323–328 (2016).
Lin, H., Long, E., Chen, W. & Liu, Y. Documenting rare disease data in China. Science 349, 1064 (2015).
Zhao, L. et al. Lanosterol reverses protein aggregation in cataracts. Nature 523, 607–611 (2015).
Lenhart, P. D. et al. Global challenges in the management of congenital cataract: proceedings of the 4th International Congenital Cataract Symposium. J. AAPOS 19, e1–e8 (2015).
Kohavi, R. in Proceedings of the 14th International Joint Conference on Artificial Intelligence 1137–1143 (2001).
Yang, J. et al. Exploiting ensemble learning for automatic cataract detection and grading. Comp. Meth. Progr. Biomed. 2016, 45–57 (2015).
Guo, L., Yang, J., Peng, L., Li, J. & Liang, Q. A computer-aided healthcare system for cataract classification and grading based on fundus image analysis. Comp. Indust. 5, 72–80 (2015).
Amaya, L., Taylor, D., Russell-Eggitt, I., Nischal, K. K. & Lengyel, D. The morphology and natural history of childhood cataracts. Surv. Ophthalmol. 48, 125–144 (2003).
Silver, D. et al. Mastering the game of Go with deep neural networks and tree search. Nature 529, 484–489 (2016).
Gulshan, V. D. et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316, 2402–2410 (2016)
Berg, A., Deng, J. & Fei-Fei, L. Large scale visual recognition challenge www.imagenet.org/challenges (accessed 16 April 2016).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Adv. Neur. Inf. Proc. Syst. 25, 2012 (2012).
Zeiler, M. D. & Fergus, R. Visualizing and understanding convolutional networks. Lecture Notes Comp. Sci. 8689, 818–833 (2013).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. Preprint at http://arxiv.org/abs/1409.1556 (2014).
LeCun, Y., Bottou, L., Bengio, Y. & Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE Inst. Electr. Electron. Eng. 86, 2278–2324 (1998).
Jarrett, K., Kavukcuoglu, K., Ranzato, M. A. & LeCun, Y. What is the best multi-stage architecture for object recognition. In Proc. IEEE 12th Int. Conf. Computer Vision 2146–2153 (2009).
Dan, C., Meier, U. & Schmidhuber, J. Multi-column deep neural networks for image classification. In Proc. IEEE 25th Conf. Computer Vision and Pattern Recognition 3642–3649 (2012).
LeCun, Y., Kavukcuoglu, K. & Farabet, C. Convolutional networks and applications in vision. In IEEE Int. Symp. Circuits and Systems 253–256 (2010).
Bottou, L. Large-scale machine learning with stochastic gradient descent. In Proc. 19th Int. Conf. Computational Satistics 177–186 (2010).
Hinton, G. E., Srivastava, N., Krizhevskym, A., Sutskever, I. & Salakhutdinov, R. R. Improving neural networks by preventing co-adaptation of feature detectors. Computer Sci. 3, 212–223 (2012).
Ciresan, D. C., Meier, U., Masci, J., Gambardella, L. M. & Schmidhuber, J. High-performance neural networks for visual object classification. Preprint at https://arxiv.org/abs/1102.0183 (2011).
Simard, P. Y., Steinkraus, D. & Platt, J. C. Best practices for convolutional neural networks applied to visual document analysis. In Proc. 7th Int. Conf. Document Analysis and Recognition 958–963 (2003).
Jia, Y. et al. Caffe: convolutional architecture for fast feature embedding. In Proc. 22nd ACM Int. Conf. Multimedia 675–678 (2014).
Acknowledgements
The authors acknowledge the involvement of the China Rare-disease Medical Intelligence (CRMI) Collaboration, which currently consists of Zhongshan Ophthalmic Centre (H.L., E.L. and Y.L.), Guangdong General Hospital (J. Zeng), Qingyuan People’s Hospital (Y. Lu) and the First Affiliated Hospital of Guangzhou University of Traditional Chinese Medicine (X. Yu). The CRMI Collaboration may be extended with more participating hospitals in the future. This study was funded by the 973 Program (2015CB964600), the NSFC (91546101), the Guangdong Provincial Natural Science Foundation for Distinguished Young Scholars of China (2014A030306030), the Youth Pearl River Scholar Funded Scheme (H.L., 2016), and the Special Program for Applied Research on Super Computation of the NSFC-Guangdong Joint Fund (the second phase).
Author information
Authors and Affiliations
Contributions
H.L., E.L., X.Liu and Y.L. designed the research; E.L., H.L., J.J., Z.Liu, X.W., L.W., Z.Lin, X.Li, J.C., J.L., Q.C., D.W. and W.C. conducted the study; E.L., H.L. and J.J. collected the data; E.L., H.L., J.J. and Y.A. analysed the data; E.L. and H.L. co-wrote the manuscript; all authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary algorithm information, figures, tables and test paper. (PDF 721 kb)
Supplementary Video 1
Instructions for using the CC-Cruiser website. (MP4 30017 kb)
Rights and permissions
About this article
Cite this article
Long, E., Lin, H., Liu, Z. et al. An artificial intelligence platform for the multihospital collaborative management of congenital cataracts. Nat Biomed Eng 1, 0024 (2017). https://doi.org/10.1038/s41551-016-0024
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41551-016-0024
This article is cited by
-
A Novel Grading System for Diffuse Chorioretinal Atrophy in Pathologic Myopia
Ophthalmology and Therapy (2024)
-
Anti-HER2 therapy response assessment for guiding treatment (de-)escalation in early HER2-positive breast cancer using a novel deep learning radiomics model
European Radiology (2024)
-
Predicting post-operative vault and optimal implantable collamer lens size using machine learning based on various ophthalmic device combinations
BioMedical Engineering OnLine (2023)
-
Multiomics, artificial intelligence, and precision medicine in perinatology
Pediatric Research (2023)
-
Multi-task hourglass network for online automatic diagnosis of developmental dysplasia of the hip
World Wide Web (2023)