Abstract
The “Iowa kindred,” a large Iowan family with autosomal-dominant Parkinson’s disease, has been followed clinically since the 1920s at the Mayo Clinic. In 2003, the genetic cause was determined to be a 1.7 Mb triplication of the alpha-synuclein genomic locus. Affected individuals present with an early-onset, severe parkinsonism-dementia syndrome. Here, we present a descendant of the Iowa kindred with novel, disease-associated non-motor findings of reduced heart rate variability, complete anosmia, and a rare skin condition called colloid milium. At autopsy, key neuropathological findings were compatible with diffuse Lewy body disease. Using high-resolution comparative genomic hybridization (CGH) array analysis to fine-map the genomic breakpoints, we observed two independent recombination events of the SNCA locus that resulted in a genomic triplication of twelve genes, including SNCA, and the disruption of two genes, HERC6 and CCSER1, at the genomic breakpoints. In conclusion, we provide further evidence that the mere two-fold overexpression of alpha-synuclein leads to a fulminant alpha-synucleinopathy with rapid progression and severe clinical and neuropathological features.
Similar content being viewed by others
Introduction
The Iowa kindred was initially described by Spellman in 1962 as a family with severe parkinsonism of autosomal-dominant inheritance.1 The family is of English and German descent with early age at onset (average 34 years, range 20–48 years). Disease progression is rapid with dementia and death occurring within 2–12 years after onset of symptoms. Neuropathology revealed severe degeneration of the substantia nigra in this kindred, widespread subcortical and cortical Lewy bodies, vacuolation of the cortex, nerve cell loss, and gliosis in the hippocampus. Neuritic pathology in cortical areas was detected by alpha-synuclein staining, which exceeded the magnitude of the most severe cases of dementia with Lewy bodies.2,3,4,5
In 2003, the underlying genetic cause was determined by Singleton and collaborators to be a triplication of the SNCA genomic locus on chromosome 4q21.6 Using quantitative PCR, the size of the triplication was initially narrowed down to a range between 1.61 and 2.04 Mb, containing 17 genes. A triplication of the SNCA gene on one allele (in addition to one copy of alpha-synuclein on the wildtype allele) causes two-fold up-regulation of the alpha-synuclein protein in the brain. This discovery demonstrated that a mere overexpression of wild-type alpha-synuclein could lead to a neurodegenerative condition very similar to Parkinson’s disease (PD). Two genetic screens in autosomal-dominant parkinsonism with dementia reported a frequency of duplications and triplications of ~1.5%.7,8 In a follow-up study, breakpoints for different families with chromosomal gains (duplications and triplications) were analyzed using SNP array technology and the Iowa kindred SNCA triplication was estimated to be ~1.7 Mb.9
Herein, we describe novel clinical, neuropathological and refined molecular genetic findings in a descendant from the Iowa kindred. We report non-motor findings such as reduced heart rate variability (HRV) and complete anosmia, and findings of a rare skin condition, colloid milium, which has not been previously reported in association with PD. Using high-resolution array comparative genomic hybridization (CGH) for copy number variant (CNV) analysis, we found that the SNCA triplication breakpoints disrupt two genes (HERC6 and CCSER1).
Results
Clinical description of presented case
Our patient presented at age 41 with rapidly progressive parkinsonism. The disease started with excessive fatigue, resting tremor in left arm, and a change in speech. MRI of the brain was unremarkable. He was treated with levodopa and his symptoms improved substantially. A year after levodopa treatment he experienced motor fluctuations. By age 43, symptoms progressed and his mental function was deteriorating quickly. At age 44, he showed typical motor complications with wearing off phenomenon, gait freezing, and peak-dose dyskinesias. He complained of bladder urgency, occasional constipation, and orthostatic hypotension. He reported vivid dreams and was “thrashing around” during his sleep, consistent with REM sleep behavior disorder.
A neurological examination at age 41 revealed moderate resting tremor and slight action tremor in his left hand and moderate rigidity bilaterally. Rapid sequential movements were moderately slow bilaterally. His posture was moderately stooped. His gait was normal with only reduced arm swing bilaterally and the patient recovered unaided from the pull test. His MOCA score was found to have dropped from 30/30 at age 41 to 8/30 at age 45. Olfactory function was tested at age 45 using the 40-item UPSIT. His score of 11/40 represents total anosmia (below 5th percentile) (Supplemental Fig. 1). We tested HRV and found extremely reduced HRV in a 5 min resting EKG. The HRV parameters measured were standard deviation of RR intervals (SDNN), percentage of consecutive RR intervals differing by more than 50 msec (pNN50), RR triangular index (RRTRI), width of Poincare plot SD1, low frequency (LF) normalized and low frequency/high frequency (LF/HF) ratio. The SNCA triplication patient had in all parameters measured lower values than any of the idiopathic PD patients tested (Supplemental Fig. 2) representing severe autonomic dysfunction. The Epworth Sleepiness Scale determined his level of daytime sleepiness to be 9/24, within the high normal range of 0-10. He demonstrated low color discrimination with a total error score of 124 on the Farnsworth-Munsell 100 Hue test (mean 68 for normal age group 40-49 yrs10).
His condition deteriorated rapidly prompting full-time care in a nursing home after only 6 years following diagnosis at age 47 and death at age 50.
Past medical history prior to onset of PD was unremarkable aside from a rare skin condition called colloid milium (Supplemental Figure 3), manifested by multiple dense dome-shaped pink and yellow papules on the dorsum of both hands and up to the elbows. This condition is thought to be caused by exposure to the sun, petroleum products, or skin bleaching creams containing hydroquinone.11,12
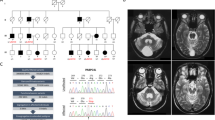
The patient is a descendent of the Iowa kindred4,5,6 and a neuropathological case report has been published on his mother. Age at onset in his mother was at 45 years with a similar rapidly progressive course including dementia.3
Neuropathology
Alpha-synuclein immunoreactive cortical Lewy bodies and Lewy neurites were most numerous in limbic and paralimbic cortices, parahippocampal cortex, insular cortex, cingulate gyrus, and in the anterior olfactory nucleus of the olfactory bulb. Additionally, there were Lewy bodies, Lewy neurites and glial inclusions in the spinal cord gray matter at the cervicomedullary junction (Fig. 1).
SNCA genomic triplication neuropathology. a Gross pathology of midbrain and pons with loss of neuromelanin pigment in both; b alpha-synuclein immunohistochemistry of locus ceruleus with Lewy bodies and bizarre neuronal inclusions; c Numerous Lewy dots in ventral tegmental region of midbrain; d spongiform change in neocortex in temporal and limbic lobes; e CA2 sector of hippocampus with Lewy neurites; f CA2 sector of hippocampus with tau in subset of Lewy neurites; g, h cortical Lewy bodies in temporal neocortex; cortical Lewy bodies and Lewy neurites in temporal neocortex; i hippocampal CA2 neurites; j amygdala Lewy bodies and neurites; k ventral tegmental area Lewy neurites (“Lewy dots”); l substantia nigra pars compacta Lewy bodies.alpha-synuclein immunohistochemistry (b, c, e, h), phospho alpha-synuclein (i, j, k, l), tau immunohistochemistry (f), hematoxylin and eosin stain (d, g). Bar in b = 20 μm (applies to c, e, f, g, h, i, j, and k); bar in d and l = 50 μm; measure bar in a is in mm
The basal nucleus of Meynert showed severe neuronal depopulation. Lewy bodies and Lewy neurites were numerous in the basal forebrain and hypothalamus. The amygdala had neuronal loss and marked gliosis and mild spongiform change with many Lewy neurites (curvilinear and dot-like) and Lewy bodies, especially in the cortical transition zone (25–45 per 20 × field) (Supplemental Table 1). α-synuclein immunohistochemistry showed many Lewy neurites, scattered glial inclusions and many spheroids in the globus pallidus. The putamen had numerous dot-like and curvilinear neurites and many cortical-type Lewy bodies as well as glial inclusions. The thalamus and subthalamic nucleus were unremarkable, but there were spare Lewy bodies and Lewy neurites in the anterior and medial nuclei.
The substantia nigra had severe neuronal loss with only sparse extraneuronal neuromelanin. There were Lewy bodies in residual neurons and many Lewy neurites, Lewy dots, and glial inclusions. Lewy bodies were present in the raphe nuclei and periaqueductal gray. The locus ceruleus had neuronal loss and gliosis with Lewy bodies. There were many Lewy bodies and neurites in the mesopontine tegmentum. A few Lewy neurites were even detected in the pontine base. The medulla was remarkable for intraneuritic Lewy bodies, Lewy neurites and neuronal loss in the dorsal motor nucleus of the vagus. There were many Lewy bodies and Lewy neurites in the medullary tegmentum. A few glial inclusions were noted in the inferior olivary nucleus. The cerebellum showed well preserved Purkinje and internal granular cell layers. There were rare glial inclusions in the cerebellar white matter with α-synuclein immunohistochemistry. The pituitary was histologically unremarkable. There was no TDP-43 pathology. Medial temporal lobe tau pathology was detected consistent with early argyrophilic grain disease.13
SNCA duplication/triplication breakpoints
To better understand the genomic structure and size of the SNCA triplication, we performed high resolution CNV analysis (1 M CGH array) to refine the breakpoints of the SNCA triplication (Fig. 2). For comparison, we also performed CNV analysis on another SNCA triplication patient (Lister kindred, Swedish–American descent),14,15 whose right breakpoint closely maps to the one found in the Iowa kindred.9 Evidence of a common founder is unlikely based on chromosome 4q21 microsatellite analysis for markers D4S3474, D4S3479, and D4S3476. The Iowa kindred has allele lengths of 185/197/205, 163/165/167, and 316/332/334,9 whereas the Lister kindred carries alleles 199/203, 163/167/169, and 332/334.9,15 The results from the 1 M CGH array (Fig. 2) clearly delineate the regions of triplication in these two cases. Interestingly, small regions of a duplication were also detected in our patient with a high-density 60 K custom CGH array (Fig. 2c), suggesting two independent mutational events. We find in our higher resolution CNV analysis that both the left and right breakpoints disrupt genes, HERC6 (left breakpoint) and CCSER1 (right breakpoint) (Fig. 2c and Supplemental Table 2). In silico analysis of the breakpoint regions revealed several repetitive elements, a LINE L2a and L2c as well as a LTR26E element, however, we did not find overt homology that would explain the rearrangement events (Supplemental Figure 5).
Chromosomal breakpoint map of SNCA triplications. a Array CGH data for the Iowa kindred case (top data track) and a patient from the Lister kindred (bottom data track). The Iowa triplicated region is 1.7 Mb and the Lister triplicated region is 0.8 Mb. Duplication and triplication breakpoints determined by the algorithm DNAcopy are indicated by red arrows. b Genome browser (UCSC, hg19) view of RefSeq genes located within and flanking the SNCA triplications. Copy numbers of 2, 3, and 4 are denoted by light, medium, and dark orange line segments for the SNCA triplication patients shown in panel (a). Vertical gray-shaded bars demarcate the breakpoints in the gene track. c Array CGH fine-mapping of Iowa kindred breakpoints: top data track is a zoomed view of the 1 M probe array data (panel a) on the Iowa male case report, middle data track is the Iowa male using a higher resolution custom CGH array, bottom data track is an Iowa female on the custom CGH array. See supplemental table 2 for genome coordinates. A Log2 ratio value of 0 corresponds to no change in copy number relative to a reference genome (see Methods), whereas duplicated and triplicated chromosomal regions have Log2 ratio values of ~0.6 and ~1.0, respectively
We confirmed the disruption of both genes by mRNA expression analysis and found decreased expression for both HERC6 and CCSER1 compared to SNCA and MMRN1 in induced pluripotent stem cells (iPSCs) from our patient differentiated into dopaminergic neurons compared to cells from a sibling control (Supplemental Figure 6). We tested expression for SNCA, MMRN1, CCSER1, as well as three HERC genes in the triplicated region, HERC3, HERC5, and HERC6. All genes in the SNCA triplication region showed an increase in mRNA expression between 2–3.5 fold compared to iPSCs from a sibling control (Supplemental Figure 6A). When we differentiated the iPSCs into a mixed population of dopaminergic neurons with ~25–35% of neurons expressing tyrosine hydroxylase (TH), the rate limiting enzyme for dopamine synthesis, we found that the SNCA and HERC3 gene expression were 2-fold increased, MMRN1 expression was ~6-fold elevated, and HERC5 mRNA expression was markedly reduced compared to the sibling control possibly due to disrupted regulatory genomic elements. Also as shown in previous work from our group, the expression of TH was significantly reduced as compared to a healthy sibling control.16
Discussion
Non-motor symptoms are becoming increasingly important and critical for early diagnosis of PD. In recent studies, sense of smell17,18,19,20,21 and cardiac autonomic denervation, which can be measured as reduction of HRV22,23 or myocardial scintigraphy (MIBG),24,25 have been used as early clinical indicators of PD.
In this report, we show that our patient with the SNCA gene triplication scores the lowest for both sense of smell on the UPSIT and has the lowest HRV values overall in a group of sporadic, typical PD. It is noteworthy that our patient presented here is also the youngest subject studied. Our patient and other published cases8,15,17,26 hint that patients with SNCA genomic triplications exhibit impaired olfaction more severe than other SNCA variants (duplications and single base mutations), possibly due to a dose effect of alpha-synuclein protein.
The other interesting clinical finding in our patient is the presentation of skin lesions, called colloid milium, a rare skin condition that has not been reported in conjunction with PD. Only one other report links this skin disorder to a genetic disease beta-thalassemia.27 The cause of colloid milium is unknown, however, high exposure to sun, petrol substances, and dermal bleaching creams are thought to have an impact on this condition.11,12
The neuropathological findings in our case are consistent with diffuse Lewy body disease with typical distribution and density of cortical Lewy bodies and severe CA2/3 neuritic pathology in addition to the severe neuronal loss in the ventrolateral cell group of the substantia nigra. Similar neuropathology has been described in other affected individuals of the Iowa kindred including the mother of our patient.3
Refined genomic breakpoint analysis of the DNA in our patient with the SNCA genomic triplication revealed that there are small regions at the left and right breakpoint of a duplication (Fig. 2c), suggesting two independent mutational events. A dynamic SNCA genomic multiplication has been reported in the Lister family complex15 where a patient with an SNCA duplication (Branch J) and a patient with a triplication (Branch I) presented with shared haplotype of the SNCA/MMRN1 region and based on an extended pedigree published in 1949, a common ancestor could be determined. However, we did not detect any evidence of a duplication in the subject of the Lister kindred (Fig. 2). Additional published duplication/triplication rearrangements are shown in Supplemental Figure 4 to illustrate the variability and size of CNVs of this region.28
Furthermore, we narrowed down the left and right genomic breakpoints of the triplication and found that the HERC6 and CCSER1 genes were disrupted. This is in contrast to a lower resolution CNV analysis of this patient that mapped the breakpoints to a region between HERC6 and HERC5 and an adjacent region outside of the CCSER1 gene.9 The impact of these novel findings is that there is now evidence that the Iowa and Lister kindreds both have a triplication breakpoint (right) that disrupts CCSER1, which may account for PD or non-PD phenotypic similarities in these patients/families. In contrast, the HERC6-disrupting breakpoint we found in our Iowa kindred case may account for phenotypic differences between these two kindreds.
Based on these new genetic findings in our patient, it is interesting to discuss if one or both gene disruptions are contributing to the patient’s clinical phenotypes reported herein. The CCSER1 gene (Coiled-Coil Serine-Rich Protein 1, also known as FAM190A) has been reported as a fragile site for genomic rearrangement in cancers, with structural defects being reported in 40% of human cancers.29 More recently, knockdown of CCSER1 was found to cause cell division defects and to interact with NDEL1, a dynein regulator that plays a role in neurodevelopment and adult neurons and may be contributing to the defective dynein-dependent axon transport noted in neurodegenerative disease like Huntington’s and PD.30 The human HERC gene family comprises 6 members, encoding 2 large and 4 small proteins, which have in common a HECT ubiquitin E3-ligase domain and an RLD domain.31 Localization of HERC3, HERC5, and HERC6 (all small HERCs) to chromosome 4 indicates recent evolution and HERC5 is the youngest member and primate-specific. Interestingly, a recent article shows that HERC5 can regulate activity of ubiquitin E3 ligase parkin by ISG15 conjugation.32
Our findings in this patient with an early-onset familial form of PD expand the non-motor clinical and neuropathological phenotype of SNCA triplication. The genetic array analysis of the SNCA triplication locus demonstrates the value of high-resolution CGH arrays for the detection of copy number variants, including discrimination of duplicated and triplicated regions and refined breakpoint mapping.
Methods
SNCA triplication cases
Genomic DNA was isolated from blood for our patient, from brain for the Lister case, and from a lymphoblastoid cell line from an additional Iowa kindred case (ND00139, NINDS Repository, Coriell Institute, Camden, NJ) for the purpose of fine-mapping the duplication and triplication breakpoints.
The study was approved by an Institutional Review Board and patients who participated in this study provided written informed consent.
Clinical assessments
A complete medical history was obtained followed by a general medical and neurological examination on the Iowa kindred patient. Five minute resting ECG was taken to assess the heart rate variability. Unified Parkinson’s Disease Rating Scale, Hoehn and Yahr stage were noted and cognitive assessment was performed by using the Montreal Cognitive Assessment (MOCA). Color vision was tested by Farnsworth-Munsell 100 Hue Test (X-rite, Grand Rapids, MI, USA), 40-item University of Pennsylvania Smell Identification Test (UPSIT, Sensonics, Inc, Haddon Heights, HJ, USA) was taken to assess olfaction. Sleeping habits were assessed by questionnaires on REM sleep behavior disorder including Epworth Sleepiness Scale.
Heart rate variability (HRV)
HRV was assessed by analyzing the normal R-R intervals of a five-minute supine waking EKG. The HRV parameters measured included the times domain parameters of standard deviation of R-R intervals (SDNN), and the percentage of consecutive RR intervals differing by more than 50 milliseconds (pNN50), the geometric parameter RR triangular index (RRTRI), the minor axis standard descriptor of the Poincaré plot (SD1), and the frequency domain parameters normalized Low Frequency (LF nu) and the ratio of LF/HF. While SDNN, RRTRI, and LF are thought to represent overall HRV influenced by both sympathetic and vagal systems, the parameters SD1 and pNN50 are considered parasympathetically dominated and LF/HF ratio indicates sympathovagal balance.
Copy number variant (CNV) analysis
A comparative genomic hybridization (CGH) microarray (Design ID 021529, Agilent Technologies, Santa Clara, CA) comprising ~1 million (1 M) oligonucleotide probes uniformly distributed across the genome (2.1 Kb median probe spacing) was used to refine the breakpoints of the SNCA triplication cases. Genomic DNA samples were dye-labeled and hybridized to the 1 M CGH array according to the manufacturer’s instructions. Genomic DNA for all samples was labeled with Cy3 dye. To accurately assess CNVs across all chromosomes, sex-matched hybridizations were performed using a healthy male reference DNA sample (Population Bio, New York, NY), which was labeled with Cy5 dye. Array experiments were performed as a service by Oxford Gene Technology (Oxford, UK). The arrays were scanned using the Agilent microarray scanner, at 2 µm resolution (16 bit) and data was extracted using Feature extraction software version 10.7.3.1, grid design file 021529_D_F_20091001 and protocol CGH_107_Sep09. CNV analysis was performed using DNAcopy, a circular binary segmentation algorithm (from the R Bioconductor package), with log2ratio cutoffs of −0.35 for losses and +0.35 for gains. Array CGH data points were also manually inspected to verify the breakpoints called by the DNAcopy algorithm.
In order to further fine-map the SNCA breakpoints, we designed a high-density 8 × 60 K custom CGH array using the Agilent eArray web portal (Agilent Technologies, Santa Clara, CA, USA). In addition to Agilent control and normalization probes, the array consisted of 52,826 probes designed to interrogate 21 genes of interest, either in their entirety or for the purpose of fine mapping of previously identified breakpoints of interest in these genes, including SNCA. The median probe spacing was 24 bp for the left breakpoint and 46 bp for the right breakpoint. The Iowa kindred male (case report) and female (Coriell ND00139) genomic DNA samples were labeled and hybridized using the methods described for the 1 M CGH array, including sex-matched co-hybridization with a healthy male or healthy female reference DNA (Population Bio, New York, NY). Arrays were scanned, data extracted (grid design file IS-62976-8-V2_8by60K_cGH_Hs_20080925), and CNV analysis was performed using the same methods described for the 1 M CGH array.
Histopathology
At the time of the autopsy the brain was divided in the sagittal plane, with the entire left hemibrain frozen at −70 °C and the right hemibrain fixed in 10% neutral buffered formalin. Multiple sections of neocortex, hippocampus, basal forebrain, basal ganglia, thalamus, midbrain, pons, medulla and cerebellum were embedded in paraffin and sections were examined with H&E microscopy. Sections of cortex, hippocampus, basal forebrain and brain stem were also stained with immunocytochemical methods and antibodies to alpha-synuclein (NACP98, Mayo Clinic, non-commercial4,33), phospho alpha-synuclein (pSyn#64, mouse monoclonal, Wako, Cat. No. 015-25191, dilution 1:10,000) and tau antibody CP13(tau phospho Ser202, mouse monoclonal, non-commercial, Peter Davies, Albert Einstein College of Medicine, Bronx, N.Y.34).
iPSCs maintenance, propagation, and dopaminergic differentiation
iPSCs from our Iowa SNCA triplication patient and sibling control35 were cultured and maintained on Geltrex (Thermo Fisher, catalog # A1413302) in Essential 8 media (Thermo Fisher, catalog # A1517001). Cells were propagated every 7 to 8 days manually without enzymatic treatment.
iPSCs were differentiated using a commercially available dopaminergic (DA) neuron differentiation kit (Thermo Fisher, catalog # A3147701) according to manufacturer’s instructions. The yield of TH neurons was 25–35% and >90% total neurons (beta-III-tubulin, TUJ1) from iPSCs after 35 days in vitro.
mRNA expression analysis
Total mRNA was isolated from iPSCs and neurons with RNeasy Micro Kit (Qiagen, catalog # 74004), following the manufacturer’s instructions. Two micrograms were used to synthesize cDNA (20 μl per reaction volume) using the iScript™ cDNA Synthesis Kit (Bio-Rad, catalog # 170-8890) in a Bio-Rad DNAEngine Peltier thermal cycler. Quantitative PCR analyzation of 1 μl of 10 ng/ul cDNA was performed in triplicates on a CFX96 Real time system Thermal cycler (Bio Rad). The primers/probes used for real-time amplification for HERC3 was FAM-MGB labeled HERC3 (Thermo Fisher, Assay ID: Hs01040150_m1), for HERC5 was FAM-MGB labeled HERC5 (Thermo Fisher, Assay ID: Hs00180943_m1), for HERC6 was FAM-MGB labeled HERC6 (Thermo Fisher, Assay ID: Hs00215555_m1), for aSyn was FAM-MGB labeled SNCA (Thermo Fisher, Assay ID: Hs00240906_m1), for MMRN1 was FAM-MGB labeled MMRN1 (Thermo Fisher, Assay ID: Hs01113299_m1), for CCSER1 was FAM-MGB labeled CCSER1 (Thermo Fisher, Assay ID: Hs00286784_m1), for TH was FAM-MGB labeled TH (Thermo Fisher, Assay ID: Hs00165941_m1), and for normalization VIC-MGB_PL labeled GAPDH (Thermo Fisher, catalog # 4326317E). Relative expression levels were calculated with subsequent ΔCT values that were analyzed using CFX software. Comparative ΔΔCT method was used to normalize with subsequent CT values to the housekeeping gene GAPDH.
Data availability
The data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Spellman, G. G. Report of familial cases of parkinsonism. Evidence of a dominant trait in a patient’s family. JAMA 179, 372–374 (1962).
Fujishiro, H. et al. Diversity of pathological features other than Lewy bodies in familial Parkinson’s disease due to SNCA mutations. Am. J. Neurodegener. Dis. 2, 266–275 (2013).
Waters, C. H. & Miller, C. A. Autosomal dominant Lewy body parkinsonism in a four-generation family. Ann. Neurol. 35, 59–64 (1994).
Gwinn-Hardy, K. et al. Distinctive neuropathology revealed by alpha-synuclein antibodies in hereditary parkinsonism and dementia linked to chromosome 4p. Acta Neuropathol. 99, 663–672 (2000).
Muenter, M. D. et al. Hereditary form of parkinsonism—dementia. Ann. Neurol. 43, 768–781 (1998).
Singleton, A. B. et al. Alpha-synuclein locus triplication causes Parkinson’s disease. Science 302, 841 (2003).
Ibanez, P. et al. Alpha-synuclein gene rearrangements in dominantly inherited parkinsonism: frequency, phenotype, and mechanisms. Arch. Neurol. 66, 102–108 (2009).
Nishioka, K. et al. Clinical heterogeneity of alpha-synuclein gene duplication in Parkinson’s disease. Ann. Neurol. 59, 298–309 (2006).
Ross, O. A. et al. Genomic investigation of alpha-synuclein multiplication and parkinsonism. Ann. Neurol. 63, 743–750 (2008).
Kinnear, P. R. & Sahraie, A. New Farnsworth–Munsell 100 hue test norms of normal observers for each year of age 5–22 and for age decades 30–70. Br. J. Ophthalmol. 86, 1408–1411 (2002).
Innocenzi, D., Barduagni, F., Cerio, R. & Wolter, M. UV-induced colloid milium. Clin. Exp. Dermatol. 18, 347–350 (1993).
Findlay, G. H., Morrison, J. G. & Simson, I. W. Exogenous ochronosis and pigmented colloid milium from hydroquinone bleaching creams. Br. J. Dermatol. 93, 613–622 (1975).
Ferrer, I., Santpere, G. & van Leeuwen, F. W. Argyrophilic grain disease. Brain 131, 1416–1432 (2008).
Farrer, M. et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann. Neurol. 55, 174–179 (2004).
Fuchs, J. et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology 68, 916–922 (2007).
Oliveira, L. M. et al. Elevated alpha-synuclein caused by SNCA gene triplication impairs neuronal differentiation and maturation in Parkinson’s patient-derived induced pluripotent stem cells. Cell Death Dis. 6, e1994 (2015).
Doty, R. L. Olfactory dysfunction in Parkinson disease. Nat. Rev. Neurol. 8, 329–339 (2012).
Doty, R. L. Olfaction in Parkinson’s disease and related disorders. Neurobiol. Dis. 46, 527–552 (2012).
Ruiz-Martinez, J. et al. Olfactory deficits and cardiac 123I-MIBG in Parkinson’s disease related to the LRRK2 R1441G and G2019S mutations. Mov. Disord. 26, 2026–2031 (2011).
Silveira-Moriyama, L. et al. Olfactory heterogeneity in LRRK2 related Parkinsonism. Mov. Disord. 25, 2879–2883 (2010).
Saunders-Pullman, R. et al. Olfactory dysfunction in LRRK2 G2019S mutation carriers. Neurology 77, 319–324 (2011).
Barbic, F. et al. Early abnormalities of vascular and cardiac autonomic control in Parkinson’s disease without orthostatic hypotension. Hypertension 49, 120–126 (2007).
Takatsu, H. et al. Cardiac sympathetic denervation from the early stage of Parkinson’s disease: clinical and experimental studies with radiolabeled MIBG. J. Nucl. Med. 41, 71–77 (2000).
Satoh, A. et al. Loss of 123I-MIBG uptake by the heart in Parkinson’s disease: assessment of cardiac sympathetic denervation and diagnostic value. J. Nucl. Med 40, 371–375 (1999).
Taki, J., Yoshita, M., Yamada, M. & Tonami, N. Significance of 123I-MIBG scintigraphy as a pathophysiological indicator in the assessment of Parkinson’s disease and related disorders: it can be a specific marker for Lewy body disease. Ann. Nucl. Med. 18, 453–461 (2004).
Kertelge, L. et al. Impaired sense of smell and color discrimination in monogenic and idiopathic Parkinson’s disease. Mov. Disord. 25, 2665–2669 (2010).
Giordano, G. et al. A case of colloid milium in patient with beta thalassaemia major. J. Cutan. Pathol. 35, 566–569 (2008).
Piper, D. A., Sastre, D. & Schüle, B. Advancing stem cell models of alpha-synuclein gene regulation in neurodegenerative disease. Front. Neurosci. https://doi.org/10.3389/fnins.2018.00199 (2018).
Glover, T. W., Wilson, T. E. & Arlt, M. F. Fragile sites in cancer: more than meets the eye. Nat. Rev. Cancer 17, 489–501 (2017).
Pandey, J. P. & Smith, D. S. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J. Neurosci. 31, 17207–17219 (2011).
Sanchez-Tena, S., Cubillos-Rojas, M., Schneider, T. & Rosa, J. L. Functional and pathological relevance of HERC family proteins: a decade later. Cell Mol. Life Sci. 73, 1955–1968 (2016).
Im, E., Yoo, L., Hyun, M., Shin, W. H. & Chung, K. C. Covalent ISG15 conjugation positively regulates the ubiquitin E3 ligase activity of parkin. Open Biol. https://doi.org/10.1098/rsob.160193 (2016).
Beach, T. G. et al. Evaluation of alpha-synuclein immunohistochemical methods used by invited experts. Acta Neuropathol. 116, 277–288 (2008).
Herskovits, A. Z. & Davies, P. The regulation of tau phosphorylation by PCTAIRE 3: implications for the pathogenesis of Alzheimer’s disease. Neurobiol. Dis. 23, 398–408 (2006).
Byers, B. et al. SNCA triplication Parkinson’s patient’s iPSC-derived DA neurons accumulate alpha-synuclein and are susceptible to oxidative stress. PloS One 6, e26159 (2011).
Acknowledgements
The patient’s motto was “I am taking one for the team!”. We are indebted to the patients and families participating in research for their commitment to help moving the discoveries towards a cure. We thank Andrew Nguyen for helping with Q-RT-PCR sample preparation. The study was supported by the Blume foundation (B.S.) and Parkinson Alliance (B.S., J.W.L.).
Author information
Authors and Affiliations
Contributions
B.S. conceived clinical and experimental study, wrote first draft of manuscript, has been responsible for study oversight, human subjects protocol, and study management. F.Z. performed expression analysis study of the breakpoint in induced pluripotent stem cells and differentiated neurons. R.A.V. performed HRV studies and analysis. S.K. coordinated and supported the clinical studies and generated figures. K.K.J. and J.W.T. did clinical assessments of patient and interpretation of clinical diagnosis. E.H. and P.S.E. performed C.G.H. experiments and analysis. A.L.S.C. did dermatological assessment and diagnosis. D.W.D. performed neuropathology and interpretation. All authors participated in writing and reviewing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.H. and P.S.E. are employees of Population Bio. The remaining authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zafar, F., Valappil, R.A., Kim, S. et al. Genetic fine-mapping of the Iowan SNCA gene triplication in a patient with Parkinson’s disease. npj Parkinson's Disease 4, 18 (2018). https://doi.org/10.1038/s41531-018-0054-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41531-018-0054-4
This article is cited by
-
Inactive S. aureus Cas9 downregulates alpha-synuclein and reduces mtDNA damage and oxidative stress levels in human stem cell model of Parkinson’s disease
Scientific Reports (2023)
-
Transcriptomic characterization of tissues from patients and subsequent pathway analyses reveal biological pathways that are implicated in spastic ataxia
Cell & Bioscience (2022)
-
SHFLD3 phenotypes caused by 17p13.3 triplication/ duplication encompassing Fingerin (BHLHA9) invariably
Orphanet Journal of Rare Diseases (2022)
-
Genetic heterogeneity on sleep disorders in Parkinson’s disease: a systematic review and meta-analysis
Translational Neurodegeneration (2022)
-
Linking the phenotype of SNCA Triplication with PET-MRI imaging pattern and alpha-synuclein CSF seeding
npj Parkinson's Disease (2022)