Abstract
Whole-genome sequencing and whole-exome sequencing have proven valuable for diagnosing inherited diseases, particularly in children. However, usage of sequencing data as a pharmacogenetic screening tool to ensure medication safety and effectiveness remains to be explored. Sixty-seven variants in 19 genes with known effects on drug response were compared between genome sequencing and targeted genotyping data for coverage and concordance in 98 pediatric patients. We used targeted genotyping data as a benchmark to assess accuracy of variant calling, and to identify copy number variations of the CYP2D6 gene. We then predicted clinical impact of these variants on drug therapy. We find genotype concordance across those panels to be > 97%. Concordance of CYP2D6 predicted phenotype between estimates of whole-genome sequencing and targeted genotyping panel were 90%; a result from a lower coverage depth or variant calling difficulties in our whole-genome sequencing data when copy number variation and/or the CYP2D6*4 haplotype were present. Importantly, 95 children had at least one clinically actionable pharmacogenetic variant. Diagnostic genomic sequencing data can be used for pre-emptive pharmacogenetic screening. However, concordance between genome-wide sequencing and target genotyping needs to be characterized for each of the pharmacologically important genes.
Similar content being viewed by others
Introduction
Over the last decade, there has been significant growth in the use of genetic information to individualize clinical care. Pharmacogenetic testing in particular has seen a surge in interest because of increased patient safety awareness programs and the opportunity to identify patients who are likely to respond to certain medications and/or those in whom there is a high probability of developing severe adverse drug reactions attributed to individual genetic variants.1, 2 However, the majority of these studies are in adult cohorts. Although pharmacogenetic pediatric research has already yielded promising results,3 advances in genome-sequencing technologies now provide the opportunity to broaden and deepen the scope of pediatric pharmacogenetics as a pre-emptive medication safety screening tool.
Currently, most laboratories conducting pharmacogenetic testing use targeted genotyping technologies to clinically screen for specific variants with well-characterized drug-gene interactions. Examples of these technologies include single or multiplexed PCR assays using Taqman hydrolysis probe chemistry (Life Technologies), mass spectrometry (Agena Biosciences), bead-based immunoassay testing (Luminex), and microarrays (Affymetrix).4, 5
In addition to providing a powerful tool for diagnosing inherited disorders in childhood,6 whole-exome sequencing (WES) and whole-genome sequencing (WGS) carry the promise to identify clinically relevant pharmacogenetic variants. Mining sequence data for pharmacogenetic variants is particularly appealing in pediatric patients as it serves as an example of predictive and individualized medicine. Yet, in order to use this data confidently, it needs to be established whether variants in pharmacogenes are adequately covered and accurately genotyped from these genome-sequencing platforms. Previous comparisons between exome/genome and targeted genotyping show potential in this area,7, 8 however, performance assessments of those platforms and estimating copy number variation (CNV) in pharmacogenes in the same pediatric patient cohort have not been explored.
Drawing from a cohort of 98 children who underwent WGS for diagnostic purposes,6 we examined the coverage of WGS along with concordance between WGS and targeted genotyping for a set of 67 single-nucleotide polymorphism (SNP) and indel variants in 19 pharmacogenes. We also compared estimates of CYP2D6 gene copy number between WGS and targeted genotyping. Although not the primary objective, we investigated the utility of WES for pharmacogenetic analysis for 12 samples in the cohort because these data were available. SNP selection was based on published drug—gene dosing guidelines as well as known drug—gene interactions with potential for future pharmacogenetic guidelines. Furthermore, we explored whether variants examined carried the potential to inform future medication decisions and thus provide an opportunity to enhance patient safety.
Results
Extraction of pharmacogenetic data from various testing platforms
To determine the accuracy of pharmacogenetic data extracted from one genomic-sequencing platform (Complete Genomics), we compared the genotype calls for 67 pharmacogenetic loci (Table 1) for 98 subjects to genotypes generated using two targeted genotyping panels. We used the targeted genotyping data as a benchmark to assess variant calling and to predict copy number status of the CYP2D6 gene. Following this, we predicted phenotype status for metabolizer genes. We genotyped 98 subjects with the iPLEX® ADME CYP2D6 Panel, analyzing 29 CYP2D6 SNP and indel variants, with a 99.8% success rate (2835 genotypes). For the remaining 38 variants (in 18 other genes), we designed a custom iPlex panel in a 2-well assay (Table 1). The 98 subjects were successfully genotyped for all 38 variants in this set, except for one position (ABCG2, rs2231137) for one sample.
Analysis of the WGS data revealed an average depth of coverage of 20X or greater for all 67 loci (average range 21.6X—79.5X: Fig. 1a) across the 98 subjects. Passing quality calls were observed for 96% of the data (6312 of the 6566 positions; Fig. 1b). This metric was over 99% in non-CYP2D6 positions. Another genomic position (CYP2C19*9, rs17884712) with low- quality calling was observed in a portion of the study samples (17/98).
a Average read depth across the 98 study samples in WGS (complete genomics) and WES (Illumina HiSeq) data. Project loci are displayed by Reference SNP and in order of genomic coordinate (within the gene), although not to scale. Variants denoted with an asterisk (*) are located in introns. b Number of samples with missing genotypes in WGS data found across the 98 study samples for each genomic coordinate. Variants denoted with an asterisk (*) are located in introns. c Relative coverage for a 6 kb window encompassing CYP2D6 gene across 98 patient samples. An average relative coverage of 1 in CYP2D6 is assigned a copy number of 2. The shared boxes (and number above them) denote the assigned CYP2D6 copy number
CYP2D6 data analysis
Seven positions in CYP2D6 were missing calls in more than 10 patients in WGS due to low-confidence calling (Fig. 1b); rs16947, a common CYP2D6 variant found in the CYP2D6*2 haplotype among others, was the most frequently missed position (60 of the 98 subjects). However, in subjects called successfully at this position, rs16947 had an average depth of coverage of approximately 50. For subjects missing a call for rs16947 and/or additional variants, the most likely CYP2D6 diplotype was manually and individually assigned by using the CYP2D6 star nomenclature9 (Table S1). However, a diplotype could not be assigned to 14 of the 98 subjects due to missing calls in key positions. Six of these 14 subjects (Patient ID: 1022, 1031, 1066, 1075, 1092, 1108) carried rs3892097 variant defining the CYP2D6*4 haplotype and seven subjects (Patient ID: 1039, 1053, 1063, 1086, 1090, 1093, and 1096) had CYP2D6 copy number gains or losses (described below). One sample (Patient ID 1093) carried a combination of both (copy number gain + CYP2D6*4).
Lastly, we looked at coding variants in the CYP2D6 gene outside of the set examined here (Table S3). We observed three synonymous and ten non-synonymous variants that were individually examined for potential effect on the activity of CYP2D6. Twelve of the 13 variants were observed in single individuals. The majority of variants, although some predicted to be deleterious, did not change the metabolizer status of CYP2D6 for the subject. However, two variants (P41L and R329L) were investigated further because of their potential to affect metabolizer status depending on the allele they occur in. Inspection of the BAM files at these coordinates suggested that the P41L variant is on the *4 allele (P34S), although this is supported by a very small number of reads that span both variants. The phase of the R329L variant could not be determined from the BAM file. These variants remain of unknown clinical significance.
Concordance of genotype calls and copy number estimation in the WGS data as compared to targeted genotyping
Between the WGS and targeted genotyping data sets, there were six discordant genotypes in three genes (one in CYP2C9, one in HLA-A and four in IFNL3) (Table S2). However, the 254 missing or poor quality calls (the majority in CYP2D6) limited a complete comparison of these platforms.
CNV in CYP2D6 is relatively common, and has been established as important accompanying information in CYP2D6 typing.10 When we examined CYP2D6-overlapping CNVs in the Complete Genomics cnvSegmentsDiploidBeta and high ConfidenceSVEventsBeta files, only one sample was flagged as having a copy number gain of CYP2D6. We instead extracted relative coverage (defined as normalized coverage level under a diploid model, a value of “1” being 2-copy) for a 6 kb window containing CYP2D6. We observed an average relative coverage deviation of more than 0.2 from the value of 1 in 25 samples (Fig. 1c), suggesting a possibility of CNV. Seven samples displayed an average relative coverage deviation of close to 0.5 (heterozygous deletion or copy number of 1), while eight samples had values of close to 1.5 (duplication or copy number of 3). One individual had an average relative coverage deviation of 1.96 (copy number of 4). A subset of nine samples was inconclusive for copy number by this method, as they exhibited intermediate values of average relative coverage (between 0.5 and 1 or between 1 and 1.5) (Fig. 1c). Interestingly, the majority of these samples (7/9) contain the CYP2D6*4 variant rs3892097 (Table S1). In heterozygous three-copy samples, we manually inspected the reference and alternative read counts at informative positions to identify the duplicated allele. We were able to determine the duplicated allele for three samples (Patient ID: 1012, 1018, 1029) in this way. The read count at these informative positions was 75 or greater in all three samples (Table S1).
The Agena Typer software identified seven samples with one copy of CYP2D6, 81 samples with two copies, nine samples with three copies and one sample with four copies of CYP2D6. However, in heterozygous three-copy samples we could not confidently deduce the duplicated allele by manual inspection of allele-specific peak heights of informative SNPs.
There were two discordant samples in the estimation of the CYP2D6 CNVs between WGS and targeted genotyping data: (1) one sample (Patient ID: 1088) was estimated to be one copy with WGS, but two copy with targeted genotyping panel and (2) one sample (Patient ID: 1075) was estimated to be two copy with WGS but three copy with the targeted genotyping panel. Of the nine samples (Patient ID: 1009, 1025, 1031, 1043, 1068, 1070, 1072, 1074, 1112) inconclusive for genomic copy number of CYP2D6 in WGS data, one sample (Patient ID: 1074) was estimated to be one copy while the other eight samples were classified as a two copy using the targeted genotyping panel (Table S1).
Concordance of genotype calls in WES as compared to WGS and targeted genotyping platforms
Data from WES were available for 12 samples from the same patient cohort and so was included in our analysis. In the 12 WES samples, the 67 loci were sequenced to an average depth of 140X, and there were no missing calls for variants in exonic regions (Fig. 1a). Two positions over 1 kb upstream of exon 1 (CYP2D6: rs28735595, rs1080985) were not covered by this WES data set, however, all other CYP2D6 positions were successfully genotyped. Considering positions with passing quality calls (n = 6312), the concordance between WGS, WES, and the targeted genotyping panel was high (> 99.9%).
Clinical utility of pharmacogenetic data using genome-wide-sequencing platforms
In order to gain insight into the potential clinical utility of pharmacogenetic data extracted from genome-wide-sequencing platforms, we merged genetic data from WGS and targeted genotyping platforms for all 98 children and used published pharmacogenetic guidelines in order to review drug-gene interactions (Table 2). In the combined data, we were able to predict phenotypes of genes involved in metabolism and elimination of medications for all 98 subjects (Fig. S1). We detected at least one clinically relevant variant in 95 of the 98 subjects that could point to an individualized drug selection and/or dosing adjustment (Fig. 2).
We considered the relevance of the pharmacogenetic information for each individual and divided the pharmacogenetic dosing guidelines into medical subspecialties. Our analyses showed that 70% of patients from this cohort would specifically benefit from dose adjustments in drugs that are mainly used in cardiology and psychiatry, followed by infectious diseases (54%), neurology (42%), gastroenterology (30%), transplant (25%), pain (9%), and oncology (8%). Assessment of the drug–gene interactions revealed that 23% of our patient samples had an increased risk of developing serious adverse events in drugs used in neurology (9%), oncology (8%), infectious disease (6%), and pain management (3%) (Fig. S2).
Our findings highlight the potential for proactive pharmacogenetics using genome-sequencing data to prevent exposing individuals to an increased risk of developing adverse drug events or therapeutic failure of medications with known drug–gene interactions over a lifetime.
Discussion
Genomic-sequencing technologies are now being translated into clinical care and have significantly improved the ability to establish diagnoses in inherited disorders. Since most of these disorders manifest in childhood, the role of genomic-sequencing technologies in pediatrics is particularly poignant. Genome sequencing carries a significant promise for the field of pharmacogenetics, an area that provides the basis to prevent severe side effects and ineffective drug treatments.11
It has previously been reported that genomic-sequencing data can be mined for pharmacogenetics variants.12, 13 Several studies of genomic-sequencing platform comparisons involving some pharmacogenetic variants have been reported.7, 8 These studies generally conclude that concordance between sequencing platforms is high for common genetic variants in coding regions. In our study, we systematically analyzed 67 SNP and indel variants with reported clinical pharmacogenetic relevance across three different platforms (WGS, WES, and targeted genotyping) in the same patient cohort. Our goal was to not only confirm that pharmacogenetic variants can be called from genomic-sequencing data sets, but also to determine (1) how well each variant was covered across samples; (2) the quality of the variant calls, and (3) the accuracy of these variants as compared to one standard method of targeted genotyping.
We observed a high concordance (> 99%) of SNP and indel variants called from WGS with those obtained from our targeted genotyping panel. However, we also found WGS was not able to accurately genotype several positions in CYP2D6. In particular, genotype information for the rs16947 SNP that defines CYP2D6*2-related haplotypes was absent in the majority of subjects (60/98) with WGS data. This required manual derivation of diplotypes based on the remaining calls and could be done with confidence in many cases. In a clinical setting, these individuals would require reflexed testing using targeted genotyping of the CYP2D6 gene. We found that individuals with an rs3892097 variant (CYP2D6*4) were more likely to have ambiguous calling (no call or only one allele confidently called) for several CYP2D6 positions. Structural variants involving the *4 haplotype are well known14, 15 and PCR-based testing for the various configurations and hybrids would be a logical follow-up in future. Our findings are also consistent with in silico modeling showing that short reads of CYP2D6 multi-align to the highly similar CYP2D7 and CYP2D8 genes,16 leading to the reasonable assumption that WGS variant calling in CYP2D6 will be platform-dependent because of variations in sequence coverage due to chemistry, read length and downstream bioinformatics tools.17 At this time, we recommend pursuing additional targeted testing for the CYP2D6 gene to confirm WGS data. Overall, six discordant genotype calls (both false positive and false negative) were observed between WGS (Complete Genomics) and our targeted genotyping panel in three different genes (CYP2C9, HLA-A, and IFNL3), which are currently being investigated (Table S2).
In our study, we identified duplications or deletions in the CYP2D6 gene in 17 out of 98 subjects, a prevalence which mirrors known CNVs of CYP2D6 in the US population.18 In three cases of copy number gain (Patient ID: 1012, 1018, 1029), we were able to manually inspect the ratios of reference and alternate allele reads at informative positions in the whole-genome data to deduce the duplicated allele. This may indicate an advantage of sequencing over genotyping, as the same could not be done confidently from the genotyping data (using peak height ratios). In five samples with a copy number gain (Patient ID: 1012, 1018, 1020, 1061, 1085) the prediction of CYP2D6 metabolizer status changed when compared to an individual of the same genotype without a duplication (Table S1). Although CYP2D6 copy number could be determined in most cases from the WGS data, copy number status was ambiguous in nine of the 98 samples examined. We speculate that mapping issues, as discussed above, complicate copy number determination. Alternatively, these individuals may harbor structural variations of CYP2D6 such as CYP2D6/CYP2D7 hybrids or complex tandem arrangements.14,15,16
Recent improvements in cost and accuracy of WES have made it feasible to use it as a molecular diagnostic tool for patients referred to evaluation of suspected genetic conditions.19 We, therefore, examined WES data (already available) from 12 individuals of our patient cohort and compared the variant calling data for the 67 positions to targeted genotyping data. Although we anticipated similar variant calling issues as observed in the WGS data, interestingly, WES variant calling in CYP2D6 was highly concordant to targeted genotyping and no position exhibited similar rates of missing data. Two SNPs outside of exons (rs28735595, rs1080985) that are traditionally examined to assign CYP2D6 haplotypes were not captured (Fig. 1a). We do note, however, that the upstream variant rs12248560 (CYP2C19*17) was covered at a sufficient depth for variant calling (average 47X) indicating that some commercial bait sets may be supplemented to capture intronic regions.20 Current algorithms for CNV detection from WES data are limited in their performance,21, 22 therefore, we did not attempt to detect CNV’s from WES data.
During the last centuries, medical practice has undergone a significant transformation. In the 19th century, the focus was mainly to treat symptoms, followed by treating diseases in the 20th century. Now, as we are at the beginning of the 21st century, focus is shifting toward predictive and pre-emptive treatments of symptoms and diseases. This allows for a shift from a late curative paradigm to an early pre-emptive one, which is becoming increasingly possible.23 Pharmacogenetics will play a critical role in this paradigm shift toward predictive and pre-emptive medicine and in order to maximize its benefit it will need to be employed in the pediatric population. Although, when a genotype–phenotype relationship is identified, the effect of developmental factors such as change of enzyme activity has to be considered, as it greatly affects drug response and tolerance in children.24 Nevertheless, genetic determinants of drug response remain stable throughout life and thus offer great promise to individualized drug therapy.25 Here we demonstrated that out of 98 samples, 95 samples harbored pharmacogenetics variants with actionable clinical results as established by pharmacotherapeutic evidence-based, peer-reviewed published guidelines from the Clinical Pharmacogenetics Implementation Consortium (CPIC), the Dutch Pharmcogenetic Working Group (DPWG) and the Food and Drug Administration (FDA).26 Our data are consistent with the results described previously.25 Furthermore, 23 samples of these individuals carried pharmacogenetic variants, which are known to have a high probability of developing serious/life threatening adverse drug events (Fig. 2; Fig. S2). These findings lend weight to the view that although there is a relatively small set of medications for which pharmacogenetics offers actionable data, future prescribing for these individuals could be optimized if genetic testing were more widely and appropriately deployed in the clinic.27 Currently, only a few pediatric pharmacogenomic test kits are commercially available and used in clinical practice.28
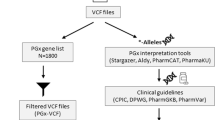
As genomic-sequencing technologies continue to improve regarding read length, data analysis and variant interpretation, pharmacogenetic testing should be considered in various primary care, outpatient and inpatient settings. We propose a two-pronged approach to the collection of pharmacogenetic data in the clinic and application to the medication prescribing process. In one arm, a conventional genotyping testing panel, for pharmacogenes with published dosing guidelines, should be made available to primary care and physician office visits, as well as hospital inpatient, outpatient, and emergency room visits as part of a laboratory blood work. In a clinical setting, where results are often required within days of administering diagnostic tests, targeted genotyping is advantageous for its cost efficiency, easy data analysis, and fast turn-around time as it directly informs applicable medication treatment choices.29 Currently, the extraction of pharmacogenes from genomic sequencing is reserved for pre-emptive information seeking individuals who are undergoing a genomic diagnostic test for an indication unrelated to pharmacogenetics. Our study provides evidence that genomic-sequencing data can also be used to extract pharmacogenetic variants. However, it is important to note that variant calling, especially in the CYP2D6 gene, could be challenging depending on sequencing platform used. For CYP2D6, manual interpretation of WGS data in the form of targeted CNV analysis, inspection of allelic read depth and decision-making surrounding missing markers, was necessary here with data generated on the Complete Genomics platform. Further PCR-based testing could also be done on some subjects to determine if they harbor structural variants of CYP2D6 not detectable by the methods here. As other WGS technologies become accessible, we recommend rigorous validation of each platform for pharmacogenetic variant calling. In whole-exome data, while most pharmacogenetics markers of interest had adequate coverage for variant calling, algorithms for copy number determination from these data are not fully developed. In the case of CYP2D6, the copy number status is an integral part of typing an individual so this testing would need to be done separately via quantitative PCR or other method. In all cases, pharmacogenetics-trained clinical pharmacists and/or pharmacologists should be involved in result interpretation and provide a report that highlights medically actionable and clinically relevant data to the primary physician, thus allowing the health-care provider to make effective and safe treatment decisions for adults as well as for pediatrics. Furthermore, it will be important to ensure that this information remain part of any electronic medical health record, improving outcomes for drug-mediated treatments over a lifetime (Fig. 3).
Workflow of incorporating PGx data into clinical care. This figure presents two clinical approaches how pre-emptive interpretation of pharmacogenetic variants can be incorporated into the medication prescribing process in the future. Pharmacogenetic data can be extracted by either a comprehensive pharmacogenetic genotyping testing panel made available to the health-care public or by genome sequencing currently used in clinical care of pediatric and adult patients. In both scenarios pharmacogenetic trained clinical pharmacists and/ or pharmacologists should be involved in assisting to interpret the results and communicate back to the ordering health-care provider and/or patient through a robust and collaborative partnership
Materials and Methods
Study cohort
Study participant and recruitment details are described elsewhere.6 Briefly, 98 children were recruited prospectively from the Genome Clinic in the Division of Clinical and Metabolic Genetics at Toronto’s Hospital for Sick Children over a 9-month period (September 2013–May 2014). DNA of the study participants was extracted from peripheral blood using the QIAsymphony DSP DNA Midi Kit on the QIAsymphony SP instrument. This study was approved by the Research Ethics Board at The Hospital for Sick Children and informed consent was obtained from all participants. Methods were performed in accordance with relevant regulations and guidelines.
Testing platforms
Whole-genome sequencing
Genomic DNA was sent to Complete Genomics (Mountain View, CA) for WGS as described previously.9 Raw sequence reads were reassembled against a reference genome (GRCh37) and variant calling was completed (and assigned a designation of ‘‘pass’’) using Complete Genomics assembly pipeline 2.4 (ref. 30). All samples passed internal Complete Genomics sample checks. Sequence results were received on hard drives and consisted of raw data plus variant calls in the form of SNP, indels, structural variants, and copy number variants. Custom VCF files were generated from the Complete Genomics files for reference and variant calls for the 67 genomic coordinates of interest (Table 1). Additional coding variants in the CYP2D6 gene outside of the set were examined from the masterVar files. Variants were filtered for passing quality. Additionally, variants which are assigned to be part of the subject’s haplotype,9 but are not genotyped in the panel, were excluded. Variants in the final list were examined for effect, if the variant is associated with a CYP2D6 allele9 and whether the variant would change the metabolizer status of the subject compared to their initial assignment. BAM files were used to confirm the phase of the variant. Variants were not validated with another method and predictions of function are theoretical only. CNV from Complete Genomics are detected through both read depth and paired-end sequencing and provided in cnvSegmentsDiploidBeta and highConfidenceSVEventsBeta files, respectively described in ref 31. We searched the cnvSegmentsDiploidBeta file for segments overlapping CYP2D6 (GRC37/hg19 chr22:42,522,501–42,526,883). Separately, we extracted relative coverage for a 6 kb region (three 2 kb windows spanning chr22:42,522,000–42,528,000).
Whole-exome sequencing
Exome capture and sequencing for 12 samples from the study cohort was performed in the Genome Diagnostics Laboratory at The Hospital for Sick Children. Briefly, exome capture was carried out using the Agilent SureSelectXT Clinical Research Exome target enrichment kit from 500 ng of genomic DNA. Sequencing (2X 100 bp paired end) was carried out on Illumina HiSeq2500 on rapid mode using V1 sequencing chemistry following the manufacturer’s instructions. Base calling was performed using CASAVA v1.8.2 and reads were mapped to the hg19 reference sequence using the BWA-backtrack algorithm from BWA v0.7.8. Duplicate reads were removed using MarkDuplicates from Picard v1.79. Local read realignment around indels, base quality score recalibration, and variant calling with GATK v2.8.1. SNP calls were subjected to variant quality score recalibration. Variant annotation was performed using annovar and custom scripts.
Targeted genotyping
All samples were genotyped for 67 positions (Table 1) using iPLEX Pro chemistry on the MassARRAY® Analyzer 4 System (Agena Biosciences, San Diego, CA, USA). Twenty-nine SNP and indel variants as well as copy number status of the CYP2D6 gene were analyzed using the Agena iPLEX® ADME CYP2D6 Panel v1.0, which is a 3-well assay combining genotyping for SNP and indel variants as well as five assays to determine genomic copy number. This genotyping platform and assay have been previously shown to accurately genotype pharmacogenetic loci in Coriell reference samples.32 CYP2D6 diplotypes and CNV calling for MassArray data were determined using the Agena PGx Report 2.0 Reporter plugin for the Typer Analyzer software (Agena Bioscience). CYP2D6 copy number was estimated from the five copy number assays that are integrated in the CYP2D6 genotyping panel, which is calculated from informative polymorphisms between CYP2D6 and CYP2D7. Variants in the remaining 18 genes in this study were also typed on the MassARRAY® Analyzer 4 System with custom-designed primers using a combination of Agena’s Assay Design Suite (ADS) and Primer3 (ref 33).
Comparison among WGS, WES, and MassArray-based targeted genotyping
Each position was manually examined for quality and read depth (in the case of sequencing) and targeted genotyping were also manually inspected. We examined genotype concordance between WGS, WES, and targeted genotyping, and CNV concordance between WGS and targeted genotyping only.
Phenotype prediction of samples based on consolidated data
The clinical utility of pharmacogenetics data was examined for all 98 samples, based on published guidelines established by the Clinical Pharmacogenetics Implementation Consortium (CPIC), Dutch Pharmacogenetic Working Group (DPWG) and in US. Food and Drug Administration (FDA) label recommendations. Information regarding the effects of allelic variation on dosing guidelines can be found at the Pharmacogenomics Knowledgebase (PharmGKB) website.26
We used the Human Cytochrome P450 Allele Nomenclature Database9 to define variant alleles in CYP2D6, CYP2C9, CYP2C19, and CYP3A5 genes and their effect on their respective CYP protein. Furthermore, phenotype assignments such as poor, intermediate, extensive, and ultrarapid metabolizers were determined by utilizing the corresponding published CPIC guidelines34,35,36,37,38,39,40 and for CYP2D6, the activity score system as described elsewhere.41 We utilized published guidelines available on the PharmGKB website to determine the influence of polymorphic variations in the remaining genes (HLA-A*31:01, HLA-B*57:01, IFNL3, SLCO1B1).42,43,44,45
Also, based on the genotype we subdivided the 98 pediatric sample cohort into three different categories by considering the significance of the variants on predicted drug metabolism and drug response: Category 1 for individuals with currently no pharmacogenetics variants of interest; Category 2 for individuals with variants that would benefit from pharmacogenetics-guided dosing; and finally Category 3 for individuals that carry pharmacogenetic variants, that are associated with developing a serious/life threatening response to a particular drug if treated with such.
Data availability statement
WGS data are deposited in the European Genome-phenome Archive (www.ebi.ac.uk/ega/) under accession number EGAS00001001623. In addition, the data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Crews, K. R., Hicks, J. K., Pui, C.-H., Relling, M. V. & Evans, W. E. Pharmacogenomics and individualized medicine: Translating science into practice. Clin. Pharmacol. Ther. 92, 467–475 (2012).
Zhou, Z. W. et al. Clinical association between pharmacogenomics and adverse drug reactions. Drugs 75, 589–631 (2015).
Stevens, A. et al. Paediatric perspective on pharmacogenomics. Pharmacogenomics 14, 1889–1905 (2013).
Thermo Fisher Scientific Inc., 2016. Available at: http://www.thermofisher.com/ca/en/home/life-science/pcr/real-time-pcr/real-time-pcr-assays/dme-analysis-using-taqman.html. Accessed September 14, 2016.
Pratt, V. M. et al. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and Association for Molecular Pathology collaborative project. J. Mol. Diagn. 12, 835–846 (2010).
Stavropoulos, D. J. et al. Whole-genome sequencing expands diagnostic utility and improves clinical management in paediatric medicine. NPJ Genom Med 1, 15012 (2016).
Chua, E. W. et al.. Cross-comparison of exome analysis, next-generation sequencing of amplicons, and the iPLEX® ADME PGx panel for pharmacogenomic profiling. Front. Pharmacol. 7 (2016). doi:10.3389/fphar.2016.00001.
Yang, W. et al. Comparison of genome sequencing and clinical genotyping for pharmacogenes. Clin. Pharmacol. Ther. 100, 380–388 (2016).
Sim, S. C. & Ingelman-Sundberg, M. Update on allele nomenclature for human cytochromes P450 (CYP) and the human cytochrome P450 allele (CYP-Allele) nomenclature database. Methods Mol. Biol. 987, 251–259 (2013).
Ramamoorthy, A. et al. Differential quantification of CYP2D6 gene copy number by four different quantitative real-time PCR assays. Pharmacogenet. Genomics 20, 451 (2010).
Swen, J. J. et al. Pharmacogenetics: from bench to byte—an update of guidelines. Clin. Pharmacol. Ther. 89, 662–673 (2011).
Kozyra, M., Ingelman-Sundberg, M. & Lauschke, V. M. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet. Med. 19, 20–29 (2016).
Fujikura, K., Ingelman-Sundberg, M. & Lauschke, V. M. Genetic variation in the human cytochrome P450 supergene family. Pharmacogenet. Genomics 25, 584–594 (2015).
Black, J. L., Walker, D. L., O’Kane, D. J. & Harmandayan, M. Frequency of undetected CYP2D6 hybrid genes in clinical samples: impact on phenotype prediction. Drug Metab. Dispos. 40, 111–119 (2012).
Gaedigk, A. et al. CYP2D7–2D6 hybrid tandems: identification of novel CYP2D6 duplication arrangements and implications for phenotype prediction. Pharmacogenomics 11, 43–53 (2010).
Twist, G. P. et al. Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences. NPJ Genom. Med. 1, 15007, doi:10.1038/npjgenmed.2015.7 (2016).
Ross, M. G. et al. Characterizing and measuring bias in sequence data. Genome Biol. 14, R51, doi:10.1186/gb-2013-14-5-r51 (2013).
Beoris, M., Wilson, J. A., Garces, J. A. & Lukowiak, A. A. CYP2D6 copy number distribution in the US population. Pharmacogen. Genomics 26, 96 (2016).
Yang, Y. et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312, 1870–1879 (2014).
Londin, E. R. et al. Performance of exome sequencing for pharmacogenomics. Per Med. 12, 109–115 (2015).
Zhao, M., Wang, Q., Wang, Q., Jia, P. & Zhao, Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics 14, 1 (2013).
Nam, J. Y. et al. Evaluation of somatic copy number estimation tools for whole-exome sequencing data. Brief Bioinform. 17, 185–192 (2016).
Zerhouni, E. A. Clinical research at a crossroads. J. Investig. Med. 54, 171–173 (2006).
Kearns, G. L. et al. Developmental pharmacology–drug disposition, action, and therapy in infants and children. N. Engl. J. Med. 349, 1157–1167 (2003).
Goh, L. L. et al. Analysis of genetic variation in CYP450 genes for clinical implementation. PLoS ONE 12, e0169233 (2017).
PharmGKB (The Pharmacogenomics Knowledgebase). Stanford: Stanford University, 2000. https://www.pharmgkb.org/view/dosing-guidelines.do?source=#.
Relling, M. V. & Evans, W. E. Pharmacogenomics in the clinic. Nature 526, 343–350 (2015).
Sing, C. W., Cheung, C. L. & Wong, I. C. Pharmacogenomics–how close/far are we to practising individualized medicine for children? Br. J. Clin. Pharmacol. 79, 419–428 (2015).
Johnson, J. A. et al. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin. Pharmacol. Ther. 92, 437 (2012).
Drmanac, R. et al.. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
Carnevali, P. et al. Computational techniques for human genome resequencing using mated gapped reads. J. Comp. Biol. 19, 279–292 (2012).
Pratt, V. M. et al. Characterization of 137 genomic DNA reference materials for 28 pharmacogenetic genes: A GeT-RM collaborative project. J. Mol. Diagn. 18, 109–123 (2016).
Untergasser, A. et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115–e115 (2012).
Scott, S. A. et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 94, 317–323 (2013).
Crews, K. R. et al. Clinical pharmacogenetics implementation consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 95, 376–382 (2014).
Hicks, J. K. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98, 127–134 (2015).
Birdwell, K. A. et al. Clinical pharmacogenetics implementation consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin. Pharmacol. Ther. 98, 19–24 (2015).
Hicks, J. K. et al. Clinical pharmacogenetics implementation consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin. Pharmacol. Ther. 93, 402–408 (2013).
Relling, M. V. et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin. Pharmacol. Ther. 93, 324–325 (2013).
Johnson, J. A. et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin. Pharmacol. Ther. 90, 625 (2011).
Gaedigk, A. et al. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 83, 234–242 (2008).
Martin, M. A. et al. Clinical pharmacogenetics implementation consortium guidelines for HLA‐B genotype and abacavir dosing: 2014 update. Clin. Pharmacol. Ther. 95, 499–500 (2014).
Muir, A. J. et al. Clinical pharmacogenetics implementation consortium (CPIC) guidelines for IFNL3 (IL28B) genotype and PEG interferon‐α–based regimens. Clin. Pharmacol. Ther. 95, 141–146 (2014).
Ramsey, L. B. et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin‐induced myopathy: 2014 update. Clin. Pharmacol. Ther. 96, 423–428 (2014).
Amstutz, U. et al. Recommendations for HLA‐B* 15: 02 and HLA‐A* 31: 01 genetic testing to reduce the risk of carbamazepine‐induced hypersensitivity reactions. Epilepsia 55, 496–506 (2014).
Acknowledgements
We thank the patients and parents who made this study possible. Technical expertize from The Centre for Applied Genomics is greatly appreciated. We thank the patients whose participation made this project possible. This study was funded by the Centre for Genetic Medicine, The Centre for Applied Genomics, The Hospital for Sick Children, Genome Canada, The University of Toronto McLaughlin Centre and by Complete Genomics. This work was undertaken at Complete Genomics and The Hospital for Sick Children.
Author information
Authors and Affiliations
Contributions
This work was undertaken at Complete Genomics and The Hospital for Sick Children. The Hospital for Sick Children Research Ethics Board approved this study: REB #1000037726. “The SickKids Genome Clinic: Developing and evaluating clinical uses of Whole-Genome Sequencing”. I.C. and T.A.P. designed the study with contributions from C.R.M., R.D.C., S.I., I.C., T.A.P. drafted the manuscript. C.R.M., D.J.S., R.Z.H., M.S.M., S.W.S., R.D.C., S.I. edited the manuscript. I.C. and T.A.P. were responsible for phenotype–genotype analysis and interpretation. I.C. was responsible for interpretation of drug response. T.A.P. analyzed and organized the mass array data and genomic sequencing data. C.R.M., R.B., P.N.R., D.J.S. contributed to sequence data analysis. P.N.R., R.B. performed the genotyping and exome sequencing experiments. R.Z.H. contributed to data collection. N.M., S.B., M.S.M. were responsible for patient recruitment. I.C. and T.A.P. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cohn, I., Paton, T.A., Marshall, C.R. et al. Genome sequencing as a platform for pharmacogenetic genotyping: a pediatric cohort study. npj Genomic Med 2, 19 (2017). https://doi.org/10.1038/s41525-017-0021-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41525-017-0021-8
This article is cited by
-
Pharmacogenetic profiling via genome sequencing in children with medical complexity
Pediatric Research (2023)
-
Pharmacogenomics: current status and future perspectives
Nature Reviews Genetics (2023)
-
Applying the Clinician-reported Genetic testing Utility InDEx (C-GUIDE) to genome sequencing: further evidence of validity
European Journal of Human Genetics (2022)
-
Precision Child Health: an Emerging Paradigm for Paediatric Quality and Safety
Current Treatment Options in Pediatrics (2020)
-
Best practices for the analytical validation of clinical whole-genome sequencing intended for the diagnosis of germline disease
npj Genomic Medicine (2020)