Abstract
The maturation of green fleshy fruit to become colourful and flavoursome is an important strategy for plant reproduction and dispersal. In tomato (Solanum lycopersicum) and many other species, fruit ripening is intimately linked to the biogenesis of chromoplasts, the plastids that are abundant in ripe fruit and specialized for the accumulation of carotenoid pigments. Chromoplasts develop from pre-existing chloroplasts in the fruit, but the mechanisms underlying this transition are poorly understood. Here, we reveal a role for the chloroplast-associated protein degradation (CHLORAD) proteolytic pathway in chromoplast differentiation. Knockdown of the plastid ubiquitin E3 ligase SP1, or its homologue SPL2, delays tomato fruit ripening, whereas overexpression of SP1 accelerates ripening, as judged by colour changes. We demonstrate that SP1 triggers broader effects on fruit ripening, including fruit softening, and gene expression and metabolism changes, by promoting the chloroplast-to-chromoplast transition. Moreover, we show that tomato SP1 and SPL2 regulate leaf senescence, revealing conserved functions of CHLORAD in plants. We conclude that SP1 homologues control plastid transitions during fruit ripening and leaf senescence by enabling reconfiguration of the plastid protein import machinery to effect proteome reorganization. The work highlights the critical role of chromoplasts in fruit ripening, and provides a theoretical basis for engineering crop improvements.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article or its Supplementary Information. Source data are provided with this paper.
Change history
13 October 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41477-021-01018-5
References
Alexander, L. & Grierson, D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 53, 2039–2055 (2002).
Klee, H. J. & Giovannoni, J. J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59 (2011).
Seymour, G. B., Ostergaard, L., Chapman, N. H., Knapp, S. & Martin, C. Fruit development and ripening. Annu. Rev. Plant Biol. 64, 219–241 (2013).
Llorente, B., D’Andrea, L. & Rodriguez-Concepcion, M. Evolutionary recycling of light signaling components in fleshy fruits: new insights on the role of pigments to monitor ripening. Front. Plant Sci. 7, 263 (2016).
Jarvis, P. & López-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802 (2013).
Sadali, N. M., Sowden, R. G., Ling, Q. & Jarvis, R. P. Differentiation of chromoplasts and other plastids in plants. Plant Cell Rep. 38, 803–818 (2019).
Barsan, C. et al. Characteristics of the tomato chromoplast revealed by proteomic analysis. J. Exp. Bot. 61, 2413–2431 (2010).
Egea, I. et al. Chromoplast differentiation: current status and perspectives. Plant Cell Physiol. 51, 1601–1611 (2010).
Li, L. & Yuan, H. Chromoplast biogenesis and carotenoid accumulation. Arch. Biochem. Biophys. 539, 102–109 (2013).
Pesaresi, P., Mizzotti, C., Colombo, M. & Masiero, S. Genetic regulation and structural changes during tomato fruit development and ripening. Front. Plant Sci. 5, 124 (2014).
Barsan, C. et al. Proteomic analysis of chloroplast-to-chromoplast transition in tomato reveals metabolic shifts coupled with disrupted thylakoid biogenesis machinery and elevated energy-production components. Plant Physiol. 160, 708–725 (2012).
Suzuki, M. et al. Plastid proteomic analysis in tomato fruit development. PLoS ONE 10, e0137266 (2015).
Szymanski, J. et al. Label-free deep shotgun proteomics reveals protein dynamics during tomato fruit tissues development. Plant J. 90, 396–417 (2017).
Dalal, M., Chinnusamy, V. & Bansal, K. C. Isolation and functional characterization of lycopene beta-cyclase (CYC-B) promoter from Solanum habrochaites. BMC Plant Biol. 10, 61 (2010).
Llorente, B. et al. Synthetic conversion of leaf chloroplasts into carotenoid-rich plastids reveals mechanistic basis of natural chromoplast development. Proc. Natl Acad. Sci. USA 117, 21796–21803 (2020).
Pech, J. C., Bouzayen, M. & Latché, A. in Fruit Ripening: Physiology, Signaling and Genomics (eds Nath, P. & Bouzayen, M.) 28–47 (CAB International, 2014).
D’Andrea, L. et al. Interference with Clp protease impairs carotenoid accumulation during tomato fruit ripening. J. Exp. Bot. 69, 1557–1568 (2018).
D’Andrea, L. & Rodriguez-Concepcion, M. Manipulation of plastidial protein quality control components as a new strategy to improve carotenoid contents in tomato fruit. Front. Plant Sci. 10, 1071 (2019).
Ling, Q., Huang, W., Baldwin, A. & Jarvis, P. Chloroplast biogenesis is regulated by direct action of the ubiquitin–proteasome system. Science 338, 655–659 (2012).
Pan, R., Satkovich, J. & Hu, J. E3 ubiquitin ligase SP1 regulates peroxisome biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 113, E7307–E7316 (2016).
Ling, Q., Li, N. & Jarvis, P. Chloroplast ubiquitin E3 ligase SP1: does it really function in peroxisomes? Plant Physiol. 175, 586–588 (2017).
Ling, Q. et al. Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363, eaav4467 (2019).
Jarvis, P. Targeting of nucleus-encoded proteins to chloroplasts in plants (Tansley Review). New Phytol. 179, 257–285 (2008).
Schnell, D. J. The TOC GTPase receptors: regulators of the fidelity, specificity and substrate profiles of the general protein import machinery of chloroplasts. Protein J. 38, 343–350 (2019).
Demarsy, E., Lakshmanan, A. M. & Kessler, F. Border control: selectivity of chloroplast protein import and regulation at the TOC-complex. Front. Plant Sci. 5, 483 (2014).
Li, H. M. & Chiu, C. C. Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61, 157–180 (2010).
Shi, L. X. & Theg, S. M. The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta 1833, 314–331 (2013).
Yan, J., Campbell, J. H., Glick, B. R., Smith, M. D. & Liang, Y. Molecular characterization and expression analysis of chloroplast protein import components in tomato (Solanum lycopersicum). PLoS ONE 9, e95088 (2014).
Lim, P. O., Kim, H. J. & Nam, H. G. Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136 (2007).
Hou, X., Zhang, W., Du, T., Kang, S. & Davies, W. J. Responses of water accumulation and solute metabolism in tomato fruit to water scarcity and implications for main fruit quality variables. J. Exp. Bot. 71, 1249–1264 (2020).
Gray, J. E., Picton, S., Giovannoni, J. J. & Grierson, D. The use of transgenic and naturally occurring mutants to understand and manipulate tomato fruit ripening. Plant Cell Environ. 17, 557–571 (1994).
López Camelo, A. F. & Gómez, P. A. Comparison of color indexes for tomato ripening. Hortic. Bras. 22, 534–537 (2004).
Batu, A. Determination of acceptable firmness and colour values of tomatoes. J. Food Eng. 61, 471–475 (2004).
Zeng, Y. et al. A comprehensive analysis of chromoplast differentiation reveals complex protein changes associated with plastoglobule biogenesis and remodeling of protein systems in sweet orange flesh. Plant Physiol. 168, 1648–1665 (2015).
Martel, C., Vrebalov, J., Tafelmeyer, P. & Giovannoni, J. J. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157, 1568–1579 (2011).
Pan, Y. et al. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 161, 1476–1485 (2013).
Rigano, M. M., Lionetti, V., Raiola, A., Bellincampi, D. & Barone, A. Pectic enzymes as potential enhancers of ascorbic acid production through the D-galacturonate pathway in Solanaceae. Plant Sci. 266, 55–63 (2018).
Chan, K. X., Phua, S. Y., Crisp, P., McQuinn, R. & Pogson, B. J. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53 (2016).
Zhao, X., Huang, J. & Chory, J. Unraveling the linkage between retrograde signaling and RNA metabolism in plants. Trends Plant Sci. 25, 141–147 (2020).
Wu, G. Z. & Bock, R. GUN control in retrograde signaling: How GENOMES UNCOUPLED proteins adjust nuclear gene expression to plastid biogenesis. Plant Cell 33, 457–474 (2021).
Chen, Y. et al. Formation and change of chloroplast-located plant metabolites in response to light conditions. Int. J. Mol. Sci. 19, 654 (2018).
Carrari, F. et al. Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 142, 1380–1396 (2006).
Ling, Q. & Jarvis, P. Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr. Biol. 25, 2527–2534 (2015).
Woo, H. R., Kim, H. J., Lim, P. O. & Nam, H. G. Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70, 347–376 (2019).
Wang, R., Angenent, G. C., Seymour, G. & de Maagd, R. A. Revisiting the role of master regulators in tomato ripening. Trends Plant Sci. 25, 291–301 (2020).
Gao, W., Liu, W., Zhao, M. & Li, W. X. NERF encodes a RING E3 ligase important for drought resistance and enhances the expression of its antisense gene NFYA5 in Arabidopsis. Nucleic Acids Res. 43, 607–617 (2015).
Teng, Y. S., Chan, P. T. & Li, H. M. Differential age-dependent import regulation by signal peptides. PLoS Biol. 10, e1001416 (2012).
Kessler, F. Chloroplast delivery by UPS. Science 338, 622–623 (2012).
Cheung, A. Y., McNellis, T. & Piekos, B. Maintenance of chloroplast components during chromoplast differentiation in the tomato mutant green flesh. Plant Physiol. 101, 1223–1229 (1993).
Dono, G. et al. Color mutations alter the biochemical composition in the San Marzano tomato fruit. Metabolites 10, 110 (2020).
Parry, C., Blonquist, J. M. Jr & Bugbee, B. In situ measurement of leaf chlorophyll concentration: analysis of the optical/absolute relationship. Plant Cell Environ. 37, 2508–2520 (2014).
Gálvez-Valdivieso, G. et al. The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21, 2143–2162 (2009).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Schwacke, R. et al. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26 (2003).
Ossowski, S., Schwab, R. & Weigel, D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53, 674–690 (2008).
Fernandez, A. I. et al. Flexible tools for gene expression and silencing in tomato. Plant Physiol. 151, 1729–1740 (2009).
Schwab, R., Ossowski, S., Riester, M., Warthmann, N. & Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133 (2006).
Karimi, M., Inze, D. & Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Chetty, V. J. et al. Evaluation of four Agrobacterium tumefaciens strains for the genetic transformation of tomato (Solanum lycopersicum L.) cultivar Micro-Tom. Plant Cell Rep. 32, 239–247 (2013).
Sun, H. J., Uchii, S., Watanabe, S. & Ezura, H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 47, 426–431 (2006).
Koornneef, M. et al. Chromosomal instability in cell- and tissue cultures of tomato haploids and diploids. Euphytica 43, 179–186 (1989).
Karimi, M., De Meyer, B. & Hilson, P. Modular cloning in plant cells. Trends Plant Sci. 10, 103–105 (2005).
Wu, F. H. et al. Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 (2009).
Kasmati, A. R., Töpel, M., Patel, R., Murtaza, G. & Jarvis, P. Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J. 66, 877–889 (2011).
Hobson, G. E., Adams, P. & Dixon, T. J. Assessing the color of tomato fruit during ripening. J. Sci. Food Agric. 34, 286–292 (1983).
Pathare, P. B., Opara, U. L. & Al-Said, F. A. Colour measurement and analysis in fresh and processed foods: a review. Food Bioproc. Tech. 6, 36–60 (2013).
Arazuri, S., Jarén, C., Arana, J. I. & de Ciriza, J. P. Influence of mechanical harvest on the physical properties of processing tomato (Lycopersicon esculentum Mill.). J. Food Eng. 80, 190–198 (2007).
Walsby-Tickle, J. et al. Anion-exchange chromatography mass spectrometry provides extensive coverage of primary metabolic pathways revealing altered metabolism in IDH1 mutant cells. Commun. Biol. 3, 247 (2020).
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L. & Fernie, A. R. Corrigendum: Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 10, 1457 (2015).
Aronsson, H. et al. Nucleotide binding and dimerization at the chloroplast pre-protein import receptor, atToc33, are not essential in vivo but do increase import efficiency. Plant J. 63, 297–311 (2010).
Faurobert, M., Pelpoir, E. & Chaib, J. Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. Methods Mol. Biol. 355, 9–14 (2007).
Kovacheva, S. et al. In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41, 412–428 (2005).
Kovacheva, S., Bédard, J., Wardle, A., Patel, R. & Jarvis, P. Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 50, 364–379 (2007).
Huang, W., Ling, Q., Bédard, J., Lilley, K. & Jarvis, P. In vivo analyses of the roles of essential Omp85-related proteins in the chloroplast outer envelope membrane. Plant Physiol. 157, 147–159 (2011).
Suorsa, M. & Aro, E. M. Expression, assembly and auxiliary functions of photosystem II oxygen-evolving proteins in higher plants. Photosynth. Res. 93, 89–100 (2007).
Andersen, B., Koch, B. & Scheller, H. V. Structural and functional analysis of the reducing side of photosystem I. Physiol. Plant. 84, 154–161 (1992).
Luo, T. et al. Distinct carotenoid and flavonoid accumulation in a spontaneous mutant of Ponkan (Citrus reticulata Blanco) results in yellowish fruit and enhanced postharvest resistance. J. Agric. Food Chem. 63, 8601–8614 (2015).
Li, W. et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE 3, e1487 (2008).
Hruz, T. et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008, 420747 (2008).
Acknowledgements
We thank E. Johnson and R. Dhaliwal for transmission electron microscopy conducted in the Sir William Dunn School of Pathology EM Facility, D. Hauton and J. McCullagh for IC–MS conducted in the Mass Spectrometry Research Facility in the Department of Chemistry, P. Bota for GC–MS conducted in the Department of Plant Sciences, P. Bota and R. Ross for technical assistance, P. Pulido for assistance with the pigment analysis, and M. R. Rodriguez Goberna for HPLC conducted at CRAG, Spain. This work was supported by a Khazanah-Oxford Centre for Islamic Studies Merdeka Scholarship to N.M.S., by Strategic Priority Research Program (Type-B; project number: XDB27020107), Chinese Academy of Sciences to Q.L., by the Spanish Agencia Estatal de Investigación (grants BIO2017-84041-P and BIO2017-90877-REDT) to M.R.-C., and by the Biotechnology and Biological Sciences Research Council (BBSRC; grants BB/H008039/1, BB/K018442/1, BB/N006372/1, BB/R005591/1, BB/R009333/1 and BB/R016984/1) to R.P.J.
Author information
Authors and Affiliations
Contributions
Q.L. and N.M.S. designed and conducted the experiments, analysed the data and wrote the manuscript. Z.S., Y. Zhou, Y. Zeng and B.H. assisted with the fruit phenotypical analyses, qRT–PCR, immunoblotting, preparation of samples for the TEM and metabolomic experiments and data analysis. M.R.-C. performed the HPLC analysis of pigments and analysed the results. R.P.J. conceived the study, supervised the work, analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The application of CHLORAD as a technology for crop improvement is covered by a patent application (no. WO2019/171091 A).
Additional information
Peer review information Nature Plants thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
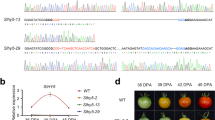
Extended Data Fig. 1 Expression profiles of the slSP1 and slSPL2 genes.
The expression profiles shown are based on Affymetrix GeneChip data and were generated using the Development (a) and Anatomy (b) functions of Genevestigator80. Data from ATH arrays are shown in scatter-plot diagrams. In a, the x-axis represents the following developmental stages, from left to right: young seedling, developed seedling, flower, green fruit, ripening fruit, and mature fruit. Values are means ± s.e.m., and for each data point the number of samples is indicated. Medium expression levels are defined as the interquartile range (IQR; light grey boxes); values below the IQR are defined as low expression (white boxes), and values above the IQR are defined as high expression (HIGH; dark grey boxes). The presented data provide a complement to the data in Fig. 1d, and confirm that slSPL2 is generally more weakly expressed than slSP1.
Extended Data Fig. 2 Assessment of the extent of knockdown or overexpression of the slSP1 and slSPL2 genes in the transgenic tomato plants.
Total leaf RNA was extracted from two-week-old tomato plants of the indicated genotypes; three independent T1 generation transformants (#1-3) were analysed for each construct. Quantitative RT–PCR analysis of slSP1 and slSPL2 expression was performed on the corresponding transgenic lines, in comparison with wild-type controls, as indicated. Relative gene expression levels were calculated by normalization using the reference gene, slACTIN. All values are expressed relative to the corresponding value for wild type, which in each case is set to 1. Values are means ± s.e.m. (n = 3 [WT-1, slSPL2-KD #2 and #3], 4 [slSP1-KD #3], or 5 [all other genotypes] technical replicates).
Extended Data Fig. 3 Determination of tomato fruit sizes.
Measurement of the maximal equatorial diameter of breaker-stage tomato fruit, from T2 generation transgenic plants, was performed using a calliper. The fruits were then detached from the plants and incubated at 25 °C in the dark for the ripening analysis presented in Fig. 3. Values are means ± s.e.m. (n = 26-27 fruits per genotype). The data demonstrate that fruit size in the slSP1-KD, slSP1-OX and slSPL2-KD transgenic lines at breaker stage was not significantly different from that in the wild type, as revealed by an unpaired two-tailed Student’s t-test (P = 0.4125 [slSP1-KD], 0.7132 [slSP1-OX], and 0.8001 [slSPL2-KD]). This rules out the possibility of nonspecific effects due to fruit size differences, which is important because ripening in detached tomato fruit is dependent on proper maturation up to the mature green stage17.
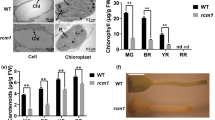
Extended Data Fig. 4 Determination of firmness of detached tomato fruits.
Fruit firmness was measured using a durometer at the breaker (Day 1) and red stages (a) (n = 20-28 [breaker stage] or 10-13 [red stage] fruits per genotype); or at specific days post breaker stage (b) (n = 20-28 [Day 1, Day 5, Day 9], 10-13 [Day 12], or 10-12 [Day 14] fruits per genotype). Note that slSP1-OX fruit at Day 14 were too soft to give a reading using the durometer. All values are means ± s.e.m. The fruit used in this analysis were randomly chosen from the fruit populations of T2 generation plants used in the ripening analysis in Fig. 3.
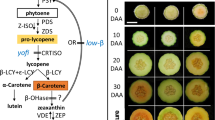
Extended Data Fig. 5 Analyses of the effects of slSP1 on ripening-related gene expression.
Total fruit RNA was extracted from wild-type (WT), slSP1-KD (KD), and slSP1-OX (OX) tomato plants at the Day 8 post-breaker stage (Day 8) stage and the red stage (the same fruit as those used in Fig. 6). Relative mRNA expression levels were analysed by qRT–PCR using primers specific for genes encoding ethylene synthesis (a), cell wall modification (b), carotenoid biosynthesis (c), and master, ripening-related transcription factors (d). It was reported previously that all of these ripening-related genes are upregulated during fruit ripening; typically, in wild-type fruit, their transcript levels will reach a peak at the pink stage, and then reduce at the red stage35,36,37. Correspondingly, in our analysis, wild-type fruit at the Day 8 post-breaker stage (pink-looking) show higher expression levels than fruit at the red stage. Although the slSP1-KD and slSP1-OX fruits both showed similar lower mRNA levels of ripening-related genes at the red stage, at Day 8 they showed striking differences in mRNA levels. In general, slSP1-KD fruit (green-looking) had markedly reduced mRNA levels, while slSP1-OX fruit (red-looking) had gene expression levels in between those of wild-type and slSP1-KD fruits. Overall, these results indicated that slSP1-KD fruit show delayed transcriptional changes of ripening-related genes relative to wild-type fruit, whereas slSP1-OX fruit displayed accelerated transcriptional changes relative to wild-type fruit. Expression data for the genes of interest were normalized using data for the reference gene, slACTIN. All values are expressed relative to the corresponding value for wild type, which in each case is set to 1. Values are means ± s.e.m. of three replicates. ACO1, 1-Aminocyclopropane-1-carboxylate oxidase 1; ACS2/4, 1-Aminocyclopropane carboxylic acid synthase 2/4; NR, Never ripe; PME, Pectin methylesterase; PG2a, Polygalacturonase 2a; PDS, Phytoene desaturase; PSY1, Phytoene synthase 1; RIN, Ripening inhibitor; TDR4, Agamous-like MADS-box protein AGL8 homolog.
Supplementary information
Supplementary Information
Supplementary Table 1.
Source data
Source Data Fig. 6
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Ling, Q., Sadali, N.M., Soufi, Z. et al. The chloroplast-associated protein degradation pathway controls chromoplast development and fruit ripening in tomato. Nat. Plants 7, 655–666 (2021). https://doi.org/10.1038/s41477-021-00916-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00916-y
This article is cited by
-
Identification and virus-induced gene silencing (VIGS) analysis of methyltransferase affecting tomato (Solanum lycopersicum) fruit ripening
Planta (2024)
-
Characterization of leucine aminopeptidase (LAP) activity in sweet pepper fruits during ripening and its inhibition by nitration and reducing events
Plant Cell Reports (2024)
-
AmCBF1 activates the expression of GhClpR1 to mediate dark-green leaves in cotton (Gossypium hirsutum)
Plant Cell Reports (2024)
-
Fruit ripening: dynamics and integrated analysis of carotenoids and anthocyanins
BMC Plant Biology (2022)
-
Plant carotenoids: recent advances and future perspectives
Molecular Horticulture (2022)