Abstract
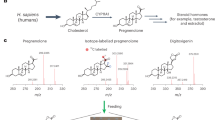
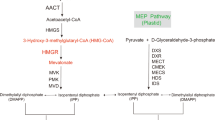
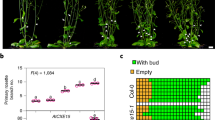
Bioactive gibberellins (GAs or diterpenes) are essential hormones in land plants that control many aspects of plant growth and development. In flowering plants, 13-OH GAs (having low bioactivity—for example, GA1) and 13-H GAs (having high bioactivity—for example, GA4) frequently coexist in the same plant. However, the identity of the native Arabidopsis thaliana 13-hydroxylase GA and its physiological functions remain unknown. Here, we report that cytochrome P450 genes (CYP72A9 and its homologues) encode active GA 13-hydroxylases in Brassicaceae. Plants overexpressing CYP72A9 exhibited semi-dwarfism, which was caused by significant reduction in GA4 levels. Biochemical assays revealed that recombinant CYP72A9 protein catalysed the conversion of 13-H GAs to the corresponding 13-OH GAs. CYP72A9 was expressed predominantly in developing seeds in Arabidopsis. Freshly harvested seeds of cyp72a9 mutants germinated more quickly than the wild type, whereas stratification-treated seeds and seeds from long-term storage did not. The evolutionary origin of GA 13-oxidases from the CYP72A subfamily was also investigated and discussed here.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data and materials generated during this study are available from the corresponding author upon request.
Change history
03 October 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Cowling, R. J., Kamiya, Y., Seto, H. & Harberd, N. P. Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol. 117, 1195–1203 (1998).
Yang, Y. Y. et al. Effects of gibberellins on seed germination of phytochrome deficient mutants of Arabidopsis thaliana. Plant Cell Physiol. 36, 1205–1211 (1995).
Magome, H. et al. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl Acad. Sci. USA 110, 1947–1952 (2013).
Blazquez, M. A., Green, R., Nilsson, O., Sussman, M. R. & Weigel, D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10, 791–800 (1998).
Eriksson, S., Bohlenius, H., Moritz, T. & Nilsson, O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18, 2172–2181 (2006).
Hedden, P. & Thomas, S. G. Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25 (2012).
Zhu, Y. Y. et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18, 442–456 (2006).
Varbanova, M. et al. Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19, 32–45 (2007).
Rieu, I. et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20, 2420–2436 (2008).
Hu, Y. L. et al. Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nat. Plants 4, 289–298 (2018).
Kanno, Y. et al. Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 51, 1988–2001 (2010).
Zhang, Y. Y. et al. Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 67, 342–353 (2011).
Nomura, T. et al. Functional analysis of Arabidopsis CYP714A1 and CYP714A2 reveals that they are distinct gibberellin modification enzymes. Plant Cell Physiol. 54, 1837–1851 (2013).
Wang, C. et al. Structural analyses of short-chain prenyltransferases identify an evolutionarily conserved GFPPS clade in Brassicaceae plants. Mol. Plant 9, 195–204 (2016).
Shao, J. et al. (+)-Thalianatriene and (-)-retigeranin B catalyzed by sesterterpene synthases from Arabidopsis thaliana. Org. Lett. 19, 1816–1819 (2017).
Chen, Q. et al. Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Sci. China Life Sci. 62, 947–958 (2019).
Jiang, C. F., Gao, X. H., Liao, L., Harberd, N. P. & Fu, X. D. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin–DELLA signaling pathway in Arabidopsis. Plant Physiol. 145, 1460–1470 (2007).
Seo, M. et al. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48, 354–366 (2006).
Finkelstein, R., Reeves, W., Ariizumi, T. & Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415 (2008).
Prall, W., Hendy, O. & Thornton, L. E. Utility of a phylogenetic perspective in structural analysis of CYP72A enzymes from flowering plants. PLoS ONE 11, e0163024 (2016).
Turk, E. M. et al. CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 133, 1643–1653 (2003).
Lange, T., Hedden, P. & Graebe, J. E. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc. Natl Acad. Sci. USA 91, 8552–8556 (1994).
Xu, Y. L. et al. The ga5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase—molecular cloning and functional expression. Proc. Natl Acad. Sci. USA 92, 6640–6644 (1995).
Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251 (2008).
Shu, K., Liu, X. D., Xie, Q. & He, Z. H. Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45 (2016).
Nelson, D. & Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 66, 194–211 (2011).
Yano, R. et al. Metabolic switching of astringent and beneficial triterpenoid saponins in soybean is achieved by a loss-of-function mutation in cytochrome P450 72A69. Plant J. 89, 527–539 (2017).
Irmler, S. et al. Indole alkaloid biosynthesis in Catharanthus roseus: new enzyme activities and identification of cytochrome P450CYP72A1 as secologanin synthase. Plant J. 24, 797–804 (2000).
Itkin, M. et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175–179 (2013).
Umemoto, N. et al. Two cytochrome P450 monooxygenases catalyze early hydroxylation steps in the potato steroid glycoalkaloid biosynthetic pathway. Plant Physiol. 171, 2458–2467 (2016).
Zi, J. C., Mafu, S. & Peters, R. J. To gibberellins and beyond! Surveying the evolution of (Di)terpenoid metabolism. Annu. Rev. Plant Biol. 65, 259–286 (2014).
Wang, G. et al. Terpene biosynthesis in glandular trichomes of hop. Plant Physiol. 148, 1254–1266 (2008).
Li, W. et al. Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell 27, 1907–1924 (2015).
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K. & Scheible, W. R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17 (2005).
Wang, G. & Pichersky, E. Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J. 49, 1020–1029 (2007).
Xu, H. et al. Characterization of the formation of branched short-chain fatty acid:CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol. Plant 6, 1301–1317 (2013).
Nelson, B. K., Cai, X. & Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 (2007).
Urban, P., Mignotte, C., Kazmaier, M., Delorme, F. & Pompon, D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 272, 19176–19186 (1997).
Pompon, D., Louerat, B., Bronine, A. & Urban, P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzym. 272, 51–64 (1996).
Martinez-Andujar, C. et al. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl Acad. Sci. USA 108, 17225–17229 (2011).
Ma, X. D. et al. CHR729 is a CHD3 protein that controls seedling development in rice. PLoS ONE 10, e0138934 (2015).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Acknowledgements
We thank X. Fu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the ga1-t mutant seeds, Q. Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the vector pCAMBIA 1300-pYAO-cas9 and J. Wu (Kunming Institute of Botany, Chinese Academy of Sciences) for the GA standard. This work was financially supported by the National Natural Science Foundation of China (grant no. 31970315), the National Key Research and Development Projects (SQ2018YFA090071-03), the ‘Priority Research Program’ of the Chinese Academy of Science (grant no. ZDRW-ZS-2019-2), the State Key Laboratory of Plant Genomics of China (SKLPG2016A-13) to G.W., the National Natural Science Foundation of China (grant no. 31770398) to J.C. and the US National Institutes of Health (grant no. GM109773) to R.J.P.
Author information
Authors and Affiliations
Contributions
G.W. designed the research. J.H. performed the majority of experiments. Q.C. and Y.M. generated the Arabidopsis transgenic plants and analysed their phenotypes. J.Y., X.W. and M.X. performed part of the biochemical assays of CYP72A members from soybean and rice. P.X. and J.C. measured the GAs in plants by ultra-high-performance liquid chromatography coupled to triple-quadrupole mass spectrometer. J.H. and G.W. analysed the data. R.J.P. and G.W. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Danièle Werck and the other, anonymous, reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–16 and Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
He, J., Chen, Q., Xin, P. et al. CYP72A enzymes catalyse 13-hydrolyzation of gibberellins. Nat. Plants 5, 1057–1065 (2019). https://doi.org/10.1038/s41477-019-0511-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0511-z
This article is cited by
-
Characterization of the horse chestnut genome reveals the evolution of aescin and aesculin biosynthesis
Nature Communications (2023)
-
Gene mining and genomics-assisted breeding empowered by the pangenome of tea plant Camellia sinensis
Nature Plants (2023)
-
CYP72D19 from Tripterygium wilfordii catalyzes C-2 hydroxylation of abietane-type diterpenoids
Plant Cell Reports (2023)
-
Cytochrome P450 CYP709C56 metabolizing mesosulfuron-methyl confers herbicide resistance in Alopecurus aequalis
Cellular and Molecular Life Sciences (2022)
-
Fine mapping and candidate gene analysis of dwarf gene Rht14 in durum wheat (Triticum durum)
Functional & Integrative Genomics (2022)