Abstract
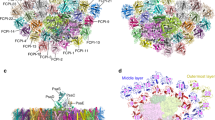
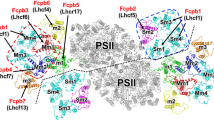
Light-harvesting antenna systems in photosynthetic organisms harvest solar energy and transfer it to the photosynthetic reaction centres to initiate charge-separation and electron-transfer reactions. Diatoms are one of the important groups of oxyphototrophs and possess fucoxanthin chlorophyll a/c-binding proteins (FCPs) as light harvesters. The organization and association pattern of FCP with the photosystem II (PSII) core are unknown. Here we solved the structure of PSII–FCPII supercomplexes isolated from a diatom, Chaetoceros gracilis, by single-particle cryoelectron microscopy. The PSII–FCPII forms a homodimer. In each monomer, two FCP homotetramers and three FCP monomers are associated with one PSII core. The structure reveals a highly complicated protein–pigment network that is different from the green-type light-harvesting apparatus. Comparing these two systems allows the identification of energy transfer and quenching pathways. These findings provide structural insights into not only excitation-energy transfer mechanisms in the diatom PSII–FCPII, but also changes of light harvesters between the red- and green-lineage oxyphototrophs during evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Atomic coordinates and Cyro-EM maps for the reported structure of C2S2, C2S1M1 and C2S2M2 have been deposited in the Protein Data Bank under accession codes 6J3Y, 6J3Z and 6J40, respectively, and in the Electron Microscopy Data Bank under accession codes EMD-9775, EMD-9776 and EMD-9777, respectively.
References
Blankenship, R. E. Molecular Mechanisms of Photosynthesis 2nd edn (Wiley-Blackwell, 2014).
Umena, Y., Kawakami, K., Shen, J.-R. & Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011).
Suga, M. et al. Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 517, 99–103 (2015).
Shen, J.-R. The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 66, 23–48 (2015).
Falkowski, P. G. et al. The evolution of modern eukaryotic phytoplankton. Science 305, 354–360 (2004).
Ballottari, M., Girardon, J., Dall’Osto, L. & Bassi, R. Evolution and functional properties of photosystem II light harvesting complexes in eukaryotes. Biochim. Biophys. Acta 1817, 143–157 (2012).
Green, B. R. in Evolution of Primary Producers in the Sea (eds Falkowski, P. G. & Knoll, A. H.) 37–53 (Elsevier Academic Press, 2007).
Engelken, J., Brinkmann, H. & Adamska, I. Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol. Biol. 10, 233 (2010).
Wei, X. et al. Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534, 69–74 (2016).
Su, X. et al. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 357, 815–820 (2017).
Liu, Z. et al. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287–292 (2004).
Standfuss, J., Terwisscha van Scheltinga, A. C., Lamborghini, M. & Kuhlbrandt, W. Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Å resolution. EMBO J. 24, 919–928 (2005).
Nagao, R. et al. Isolation and characterization of oxygen-evolving thylakoid membranes and Photosystem II particles from a marine diatom Chaetoceros gracilis. Biochim. Biophys. Acta 1767, 1353–1362 (2007).
Nagao, R. et al. Purification and characterization of a stable oxygen-evolving photosystem II complex from a marine centric diatom, Chaetoceros gracilis. Biochim. Biophys. Acta 1797, 160–166 (2010).
Nagao, R., Yokono, M., Akimoto, S. & Tomo, T. High excitation energy quenching in fucoxanthin chlorophyll a/c-binding protein complexes from the diatom Chaetoceros gracilis. J. Phys. Chem. B 117, 6888–6895 (2013).
Nagao, R., Yokono, M., Teshigahara, A., Akimoto, S. & Tomo, T. Light-harvesting ability of the fucoxanthin chlorophyll a/c-binding protein associated with photosystem II from the diatom Chaetoceros gracilis as revealed by picosecond time-resolved fluorescence spectroscopy. J. Phys. Chem. B 118, 5093–5100 (2014).
Nagao, R., Yokono, M., Tomo, T. & Akimoto, S. Control mechanism of excitation energy transfer in a complex consisting of photosystem II and fucoxanthin chlorophyll a/c-binding protein. J. Phys. Chem. Lett. 5, 2983–2987 (2014).
Ago, H. et al. Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J. Biol. Chem. 291, 5676–5687 (2016).
Okumura, A. et al. A novel protein in photosystem II of a diatom Chaetoceros gracilis is one of the extrinsic proteins located on lumenal side and directly associates with PSII core components. Biochim. Biophys. Acta 1777, 1545–1551 (2008).
Nagao, R. et al. Binding and functional properties of five extrinsic proteins in oxygen-evolving photosystem II from a marine centric diatom, Chaetoceros gracilis. J. Biol. Chem. 285, 29191–29199 (2010).
Nagao, R. et al. Crystal structure of Psb31, a novel extrinsic protein of photosystem II from a marine centric diatom and implications for its binding and function. Biochemistry 52, 6646–6652 (2013).
Nagao, R. et al. Electrostatic interaction of positive charges on the surface of Psb31 with photosystem II in the diatom Chaetoceros gracilis. Biochim. Biophys. Acta 1858, 779–785 (2017).
Nagao, R., Suzuki, T., Dohmae, N., Shen, J.-R. & Tomo, T. Functional role of Lys residues of Psb31 in electrostatic interactions with diatom photosystem II. FEBS Lett. 591, 3259–3264 (2017).
Wang, W. et al. Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science 363, eaav0365 (2019).
Akimoto, S. et al. Excitation relaxation dynamics and energy transfer in fucoxanthin-chlorophyll a/c-protein complexes, probed by time-resolved fluorescence. Biochim. Biophys. Acta 1837, 1514–1521 (2014).
Müller, P., Li, X.-P. & Niyogi, K. K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125, 1558–1566 (2001).
Akimoto, S. et al. Ultrafast excitation relaxation dynamics of lutein in solution and in the light-harvesting complexes II isolated from Arabidopsis thaliana. J. Phys. Chem. B 109, 12612–12619 (2005).
Ikeuchi, M. & Inoue, Y. A new photosystem II reaction center component (4.8 kDa protein) encoded by chloroplast genome. FEBS Lett. 241, 99–104 (1988).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Grigorieff, N. & Harrison, S. C. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr. Opin. Struct. Biol. 21, 265–273 (2011).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of coot. Acta Crystallogr. D 66, 486–501 (2010).
Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Adams, P. D. et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Barad, B. A. et al. EMRinger: side chain directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Acknowledgements
This work was supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant No. JP18am0101072j0001 (to N.M.), PRESTO from JST Grant No. JPMJPR16P1 (to F.A.), JSPS KAKENHI Nos. JP17K07442 (to R.N.), JP16H06553 (to S.A.) and JP17H06433 (to J.-R.S.), Advanced Low Carbon Technology Research and Development Program from the Japan Science and Technology Agency Grant No. JPMJAL1105 (to Y.K. and K.I.) and a Collaborative Research Program from National Institute for Basic Biology Grant No. 18-451 (to K.I.). We thank H. Nishide, NIBB, for supporting the genome data analysis.
Author information
Authors and Affiliations
Contributions
R.N., J.-R.S., N.M. and F.A. conceived the project. R.N. purified the PSII–FCPII supercomplexes and performed their biochemical and spectroscopic characterizations. T.S. and N.D. identified gene products of FCP by MS analyses. K.I, I.U. and Y.K. provided genome information of C. gracilis. N.M. collected cryo-EM images. R.N., F.A. and N.M. processed the EM data. N.M. reconstructed the final EM maps. R.N., F.A., K.K. and N.M. built the structure model. K.K. refined the final models. F.A. analysed the structure. R.N. and S.A. proposed structural interpretation on the basis of spectroscopic analyses. R.N., F.A., K.K., S.A., N.M. and J.-R.S. wrote the paper. All of the authors contributed to the interpretations of the results and improvement of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature Plants thanks Douglas Campbell and Paul Jensen and other, anonymous, reviewers for their contribution to the peer review of this work
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and Supplementary Tables 1–5.
Rights and permissions
About this article
Cite this article
Nagao, R., Kato, K., Suzuki, T. et al. Structural basis for energy harvesting and dissipation in a diatom PSII–FCPII supercomplex. Nat. Plants 5, 890–901 (2019). https://doi.org/10.1038/s41477-019-0477-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0477-x
This article is cited by
-
Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120
Nature Communications (2023)
-
Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana
Nature Communications (2023)
-
Atomic force microscopic analysis of the light-harvesting complex 2 from purple photosynthetic bacterium Thermochromatium tepidum
Photosynthesis Research (2023)
-
Solar energy conversion by photosystem II: principles and structures
Photosynthesis Research (2023)
-
Structural basis for different types of hetero-tetrameric light-harvesting complexes in a diatom PSII-FCPII supercomplex
Nature Communications (2022)