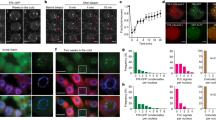
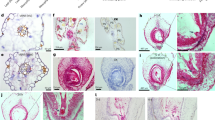
Abstract
Some overwintering plants acquire competence to flower, after experiencing prolonged cold in winter, through a process termed vernalization. In the crucifer plant Arabidopsis thaliana, prolonged cold induces chromatin-mediated silencing of the potent floral repressor FLOWERING LOCUS C (FLC) by Polycomb proteins. This vernalized state is epigenetically maintained or ‘memorized’ in warm rendering plants competent to flower in spring, but is reset in the next generation. Here, we show that in early embryogenesis, two homologous B3 domain transcription factors LEAFY COTYLEDON 2 (LEC2) and FUSCA3 (FUS3) compete against two repressive B3-containing epigenome readers and Polycomb partners known as VAL1 and VAL2 for the cis-regulatory cold memory element (CME) of FLC to disrupt Polycomb silencing. Consistently, crystal structures of B3–CME complexes show that B3FUS3, B3LEC2 and B3VAL1 employ a nearly identical binding interface for CME. We further found that LEC2 and FUS3 recruit the scaffold protein FRIGIDA in association with active chromatin modifiers to establish an active chromatin state at FLC, which results in resetting of the silenced FLC to active and erasing the epigenetic parental memory of winter cold in early embryos. Following embryo development, LEC2 and FUS3 are developmentally silenced throughout post-embryonic stages, enabling VALs to bind to the CME again at seedling stages at which plants experience winter cold. Our findings illustrate how overwintering crucifer annuals or biennials in temperate climates employ a subfamily of B3 domain proteins to switch on, off and on again the expression of a key flowering gene in the embryo-to-plant-to-embryo cycle, and thus to synchronize growth and development with seasonal temperature changes in their life cycles.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available within the paper and its Supplementary Information or from the corresponding authors on request. The coordinates and structure factors of the B3–CME complexes have been deposited in the RCSB Protein Data Bank with the accession codes: B3LEC2–DNACME: 6J9C; B3FUS3–DNACME: 6J9B; and B3VAL1–DNACME: 6J9A.
References
Tang, W. W., Kobayashi, T., Irie, N., Dietmann, S. & Surani, M. A. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 17, 585–600 (2016).
Baroux, C., Raissig, M. T. & Grossniklaus, U. Epigenetic regulation and reprogramming during gamete formation in plants. Curr. Opin. Genet. Dev. 21, 124–133 (2011).
Tao, Z. et al. Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551, 124–128 (2017).
Lamke, J. & Baurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18, 124 (2017).
Kinoshita, T. & Seki, M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 55, 1859–1863 (2014).
Andres, F. & Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639 (2012).
Sharma, N. et al. A FLOWERING LOCUS C homolog is a vernalization-regulated repressor in Brachypodium and is cold regulated in wheat. Plant Physiol. 173, 1301–1315 (2017).
Bouche, F., Woods, D. P. & Amasino, R. M. Winter memory throughout the plant kingdom: different paths to flowering. Plant Physiol. 173, 27–35 (2017).
Crevillen, P. et al. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590 (2014).
Choi, J. et al. Resetting and regulation of FLOWERING LOCUS C expression during Arabidopsis reproductive development. Plant J. 57, 918–931 (2009).
Michaels, S. D. & Amasino, R. M. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941 (2001).
Choi, K. et al. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289–303 (2011).
Yang, H. et al. Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis. Science 357, 1142–1145 (2017).
Sung, S. & Amasino, R. M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164 (2004).
Heo, J. B. & Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79 (2011).
Jiang, D. & Berger, F. DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 357, 1146–1149 (2017).
Questa, J. I., Song, J., Geraldo, N., An, H. & Dean, C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 353, 485–488 (2016).
Yuan, W. et al. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 48, 1527–1534 (2016).
Chen, N., Veerappan, V., Abdelmageed, H., Kang, M. & Allen, R. D. HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. Plant Cell 30, 600–619 (2018).
Sheldon, C. C. et al. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl Acad. Sci. USA 105, 2214–2219 (2008).
Swaminathan, K., Peterson, K. & Jack, T. The plant B3 superfamily. Trends Plant Sci. 13, 647–655 (2008).
Wang, F. & Perry, S. E. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161, 1251–1264 (2013).
Braybrook, S. A. et al. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl Acad. Sci. USA 103, 3468–3473 (2006).
Le, B. H. et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl Acad. Sci. USA 107, 8063–8070 (2010).
Yang, C. et al. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23, 1324–1329 (2013).
Suzuki, M., Wang, H. H. & McCarty, D. R. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143, 902–911 (2007).
Lee, I., Michaels, S. D., Masshardt, A. S. & Amasino, R. M. The late-flowering phenotype of FRIGIDA and LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 6, 903–909 (1994).
Pelletier, J. M. et al. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl Acad. Sci. USA 114, E6710–E6719 (2017).
Santos-Mendoza, M. et al. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54, 608–620 (2008).
Craft, J. et al. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 41, 899–918 (2005).
Boer, D. R. et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589 (2014).
Yamasaki, K. et al. Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1. Plant Cell 16, 3448–3459 (2004).
Sasnauskas, G., Kauneckaite, K. & Siksnys, V. Structural basis of DNA target recognition by the B3 domain of Arabidopsis epigenome reader VAL1. Nucleic Acids Res. 46, 4316–4324 (2018).
Wu, B. et al. Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 501, 415–422 (2018).
Ko, J. H. et al. Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J. 29, 3208–3215 (2010).
Li, Z., Jiang, D. & He, Y. FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat. Plants 4, 836–846 (2018).
Schneider, A. et al. Potential targets of VIVIPAROUS1/ABI3-LIKE1 (VAL1) repression in developing Arabidopsis thaliana embryos. Plant J. 85, 305–319 (2016).
Rosa, S., Duncan, S. & Dean, C. Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced gene repression. Nat. Commun. 7, 13031 (2016).
Schonrock, N. et al. Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 20, 1667–1678 (2006).
Michaels, S. D. & Amasino, R. M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999).
Jefferson, R. A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405 (1987).
Wang, Y., Gu, X., Yuan, W., Schmitz, R. J. & He, Y. Photoperiodic control of the floral transition through a distinct Polycomb repressive complex. Dev. Cell 28, 727–736 (2014).
Luo, X. et al. The NUCLEAR FACTOR-CONSTANS complex antagonizes Polycomb repression to de-repress FLOWERING LOCUS T expression in response to inductive long days in Arabidopsis. Plant J. 95, 17–29 (2018).
Olsen, L. J. et al. Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell 5, 941–952 (1993).
Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Karimi, M., De Meyer, B. & Hilson, P. Modular cloning in plant cells. Trends Plant Sci. 10, 103–105 (2005).
Hajdukiewicz, P., Svab, Z. & Maliga, P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994 (1994).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Di Tommaso, P. et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 (2011).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W224 (2014).
Acknowledgements
We are very grateful to R. M. Amasino for providing the FRI-Col and FRI flc seeds. We thank T. Chen and Z. Gao for anti-CLF antibody validation and the staff members at the beamline BL19U1 of the National Center for Protein Sciences Shanghai (NCPSS) at the Shanghai Synchrotron Radiation Facility (SSRF) for data collection, the staff members at the Core Facility for Molecular Biology, Shanghai Institute of Biochemistry and Cell Biology, CAS, for the help of the Octet-based in vitro binding assay. This work was supported in part by the National Key Research and Development Program of China (2016YFA0503200 to J.D. and 2017YFA0503803 to Y.H.), the National Natural Science Foundation of China (31830049 to Y.H. and 31622032 to J.D.) and the Chinese Academy of Sciences (XDB27030202 to Y.H.).
Author information
Authors and Affiliations
Contributions
Y.H. and J.D. designed and supervised the research. H.H. and B.J. performed the structure-related analyses and the in vitro protein–DNA binding assays. Z.T. and X.L. conducted the other experiments. All authors took part in data analysis. Y.H. and J.D. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information Nature Plants thanks Richard Amasino and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–8 and Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Tao, Z., Hu, H., Luo, X. et al. Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nat. Plants 5, 424–435 (2019). https://doi.org/10.1038/s41477-019-0402-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0402-3
This article is cited by
-
Application of genome editing techniques to regulate gene expression in crops
BMC Plant Biology (2024)
-
A molecular mechanism for embryonic resetting of winter memory and restoration of winter annual growth habit in wheat
Nature Plants (2024)
-
RWP-RK Domain 3 (OsRKD3) induces somatic embryogenesis in black rice
BMC Plant Biology (2023)
-
Polycomb Repressive Complexes and Their Roles in Plant Developmental Programs, Particularly Floral Transition
Journal of Plant Biology (2023)
-
A robust mechanism for resetting juvenility during each generation in Arabidopsis
Nature Plants (2022)