Abstract
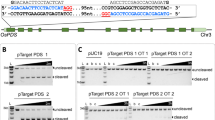
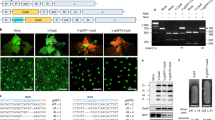
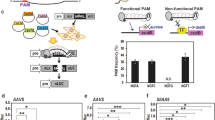
Technology involving the targeted mutagenesis of plants using programmable nucleases has been developing rapidly and has enormous potential in next-generation plant breeding. Notably, the clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein-9 nuclease (Cas9) (CRISPR–Cas9) system has paved the way for the development of rapid and cost-effective procedures to create new mutant populations in plants1,2. Although genome-edited plants from multiple species have been produced successfully using a method in which a Cas9–guide RNA (gRNA) expression cassette and selectable marker are integrated into the genomic DNA by Agrobacterium tumefaciens-mediated transformation or particle bombardment3, CRISPR–Cas9 integration increases the chance of off-target modifications4, and foreign DNA sequences cause legislative concerns about genetically modified organisms5. Therefore, DNA-free genome editing has been developed, involving the delivery of preassembled Cas9–gRNA ribonucleoproteins (RNPs) into protoplasts derived from somatic tissues by polyethylene glycol–calcium (PEG–Ca2+)-mediated transfection in tobacco, Arabidopsis, lettuce, rice6, Petunia7, grapevine, apple8 and potato9, or into embryo cells by biolistic bombardment in maize10 and wheat11. However, the isolation and culture of protoplasts is not feasible in most plant species and the frequency of obtaining genome-edited plants through biolistic bombardment is relatively low. Here, we report a genome-editing system via direct delivery of Cas9–gRNA RNPs into plant zygotes. Cas9–gRNA RNPs were transfected into rice zygotes produced by in vitro fertilization of isolated gametes12 and the zygotes were cultured into mature plants in the absence of selection agents, resulting in the regeneration of rice plants with targeted mutations in around 14–64% of plants. This efficient plant-genome-editing system has enormous potential for the improvement of rice as well as other important crop species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Information on the multiplex vector (pMgPoef4_129-2A-GFP) has been deposited in GenBank with the accession number LC460477. The authors declare that all other data supporting the findings of this study are available in the paper and its supplementary information files or from the corresponding author upon reasonable request.
References
Belhaj, K., Chaparro-Garcia, A., Kamoun, S. & Nekrasov, V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9, 39 (2013).
Voytas, D. F. Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350 (2013).
Kumar, V. & Jain, M. The CRISPR–Cas system for plant genome editing: advances and opportunities. J. Exp. Bot. 66, 47–57 (2015).
Lawrenson, T. et al. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16, 258 (2015).
Jones, H. D. Regulatory uncertainty over genome editing. Nat. Plants 1, 14011 (2015).
Woo, J. W. et al. DNA-free genome editing in plants with preassembled CRISPR–Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164 (2015).
Subburaj, S. et al. Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 35, 1535–1544 (2016).
Malnoy, M. et al. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904 (2016).
Andersson, M. et al. Genome editing in potato via CRISPR–Cas9 ribonucleoprotein delivery. Physiol. Plant. 164, 378–384 (2018).
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K. & Mark Cigan, A. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274 (2016).
Liang et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261 (2017).
Uchiumi, T., Uemura, I. & Okamoto, T. Establishment of an in vitro fertilization system in rice (Oryza sativa L.). Planta 226, 581–589 (2007).
Hwang, W. Y. et al. Efficient genome editing in zebrafish using a CRISPR–Cas system. Nat. Biotechnol. 31, 227–229 (2013).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Russell, S. D. Double fertilization. Int. Rev. Cytol. 140, 357–388 (1992).
Raghavan, V. Some reflections on double fertilization, from its discovery to the present. New Phytol. 159, 565–583 (2003).
Koiso, N., Toda, E., Ichikawa, M., Kato, N. & Okamoto, T. Development of gene expression system in egg cells and zygotes isolated from rice and maize. Plant Direct 1, e00010 (2017).
Crossway, A. et al. Micromanipulation techniques in plant biotechnology. Biotechniques 4, 320–334 (1986).
Davey, M. R., Cocking, E. C., Freeman, J., Pearce, N. & Tudor, I. Transformation of Petunia protoplasts by isolated Agrobacterium plasmids. Plant Sci. Lett. 18, 307–313 (1980).
Lurquin, P. F. Entrapment of plasmid DNA by liposomes and their interactions with plant protoplasts. Nucleic Acids Res. 6, 3773–3784 (1979).
Shillito, R. D., Saul, M. W., Paszkowski, J., Müller, M. & Potrykus, I. High efficiency direct gene transfer to plants. Nat. Biotechnol. 3, 1099–1103 (1985).
Kranz, E., von Wiegen, P. & Lörz, H. Early cytological events after induction of cell division in egg cells and zygote development following in vitro fertilization with angiosperm gametes. Plant J. 8, 9–23 (1995).
Toda, E., Ohnishi, Y. & Okamoto, T. Development of polyspermic rice zygotes. Plant Physiol. 171, 206–214 (2016).
Mikami, M., Toki, S. & Endo, M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 88, 561–572 (2015).
Jensen, K. T. et al. Chromatin accessibility and guide sequence secondary structure affect CRISPR–Cas9 gene editing efficiency. FEBS Lett. 591, 1892–1901 (2017).
Kranz, E. & Lörz, H. In vitro fertilization with isolated, single gametes results in zygotic embryogenesis and fertile maize plants. Plant Cell 5, 739–746 (1993).
Kovács, M., Barnabás, B. & Kranz, E. Electro-fused isolated wheat (Triticum aestivum L.) gametes develop into multicellular structures. Plant Cell Rep. 15, 178–180 (1995).
Kumlehn, J., Lörz, H. & Kranz, E. Differentiation of isolated wheat zygotes into embryos and normal plants. Planta 205, 327–333 (1998).
Leduc, N. et al. Isolated maize zygotes mimic in vivo embryonic development and express microinjected genes when cultured in vitro. Dev. Biol. 177, 190–203 (1996).
Holm, P. B. et al. Regeneration of fertile barley plants from mechanically isolated protoplasts of the fertilized egg cell. Plant Cell 6, 531–543 (1994).
Uchiumi, T., Komatsu, S., Koshiba, T. & Okamoto, T. Isolation of gametes and central cells from Oryza sativa L. Sex Plant Reprod. 19, 37–45 (2006).
Toda, E., Ohnishi, Y. & Okamoto, T. Electro-fusion of gametes and subsequent culture of zygotes in rice. Bio-protocol 6, e2074 (2016).
Toki, S. et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47, 969–976 (2006).
Osakabe, Y. et al. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 6, 26685 (2016).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Acknowledgements
We thank A. Ide (RIKEN Cluster for Science) for performing part of the molecular experiments, the RIKEN Bio Resource Center (Tsukuba) for providing cultured rice cells (Oc line), and the RIKEN CSRS Sequencing Service (Yokohama) for sequencing. This work was supported, in part, by the MEXT KAKENHI (Grant-in-Aid for Scientific Research on Innovative Areas, grant no. 17H05845 to T.O.) and JSPS KAKENHI (Grant-in-Aid for Challenging Exploratory Research, grant no. 16K14742 to T.O.). This work was supported by JST’s Program on Open Innovation Platform with Enterprises, Research Institute and Academia (OPERA) (Y.O.).
Author information
Authors and Affiliations
Contributions
E.T., T.K., N.Kato and T.O. designed the experiments; E.T. performed most of the experiments; N.Koiso provided technical assistance to E.T. and performed some parts of the experiments; A.T. performed some parts of the molecular experiments; M.I. prepared transformed rice plants; T.K. provided technical assistance; K.O. and Y.O. performed vector construction; H.S., N.Kato and T.O. supervised the project; and E.T. and T.O. conceived the project and wrote the article.
Corresponding authors
Ethics declarations
Competing interests
T.O., N.Kato, E.T., N.Koiso, M.I. and T.K. are co-inventors on a patent application covering the results described in this paper.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Figures 1–11, Supplementary Tables 1–10 and Supplementary References.
Rights and permissions
About this article
Cite this article
Toda, E., Koiso, N., Takebayashi, A. et al. An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants 5, 363–368 (2019). https://doi.org/10.1038/s41477-019-0386-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0386-z
This article is cited by
-
DNA-free genome editing in tomato protoplasts using CRISPR/Cas9 ribonucleoprotein delivery
Horticulture, Environment, and Biotechnology (2024)
-
Molecular insights and omics-based understanding of plant–microbe interactions under drought stress
World Journal of Microbiology and Biotechnology (2024)
-
The sonication-assisted whisker method enables CRISPR-Cas9 ribonucleoprotein delivery to induce genome editing in rice
Scientific Reports (2023)
-
Heritable transgene-free genome editing in plants by grafting of wild-type shoots to transgenic donor rootstocks
Nature Biotechnology (2023)
-
Engineering apomixis in crops
Theoretical and Applied Genetics (2023)