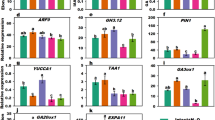
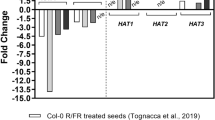
Abstract
The plant hormone gibberellin plays key roles in almost all aspects of plant development, but its detailed function and underlying regulatory mechanism in embryo development are not yet clearly defined. Here, we illustrate an essential role of gibberellin in late embryogenesis of Arabidopsis. Bioactive gibberellins are highly biosynthesized during the late developmental stage of embryos. At that time, deficiency in gibberellin biosynthesis or signalling results in an abnormal embryo phenotype characterized by less-developed cotyledons and shortened embryo axis. In contrast, gibberellin overdose leads to a significantly larger size of mature embryo. We reveal that the gibberellin signalling repressor DELLA interact with LEAFY COTYLEDON1 (LEC1), the key regulator in late embryogenesis. Gibberellin triggers the degradation of DELLAs to relieve their repression of LEC1, thus promoting auxin accumulation to facilitate embryo development. Therefore, we uncover a space/time-specific role of gibberellin in regulating late embryogenesis through the gibberellin–DELLA–LEC1 signalling cascade, providing a novel mechanistic understanding of how phytohormones regulate embryogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Goldberg, R. B., de Paiva, G. & Yadegari, R. Plant embryogenesis: Zygote to seed. Science 266, 605–614 (1994).
Cheng, Y., Dai, X. & Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439 (2007).
Luerssen, H., Kirik, V., Herrmann, P. & Misera, S. FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15, 755–764 (1998).
Stone, S. L. et al. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl Acad. Sci. USA 98, 11806–11811 (2001).
Lee, H., Fischer, R. L., Goldberg, R. B. & Harada, J. J. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl Acad. Sci. USA 100, 2152–2156 (2003).
Braybrook, S. A. & Harada, J. J. LECs go crazy in embryo development. Trends Plant Sci. 13, 624–30 (2008).
Meinke, D. W., Franzmann, L. H., Nickle, T. C. & Yeung, E. C. Leafy cotyledon mutants of Arabidopsis. Plant Cell 6, 1049–1064 (1994).
Kwong, R. W. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15, 5–18 (2002).
Lotan, T. et al. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205 (1998).
Junker, A. et al. Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71, 427–442 (2012).
Sun, T. P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 6, e0103 (2008).
Hays, D. B., Yeung, E. C. & Pharis, R. P. The role of gibberellins in embryo axis development. J. Exp. Bot. 53, 1747–1751 (2002).
Swain, S. M., Reid, J. B. & Kamiya, Y. Gibberellins are required for embryo growth and seed development in pea. Plant J. 12, 1329–1338 (1997).
Singh, D. P., Jermakow, A. M. & Swain, S. M. Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 14, 3133–3147 (2002).
Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251 (2008).
Murase, K., Hirano, Y., Sun, T. P. & Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 (2008).
Ueguchi-Tanaka, M. et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 (2005).
Lee, S. et al. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes. Dev. 16, 646–658 (2002).
Peng, J. et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes. Dev. 11, 3194–3205 (1997).
Silverstone, A. L., Ciampaglio, C. N. & Sun, T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 1555–1566 (1998).
Daviere, J. M. & Achard, P. A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 9, 10–20 (2016).
Cheng, H. et al. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064 (2004).
Li, D., Guo, Z. & Chen, Y. Direct derivatization and quantitation of ultra-trace gibberellins in sub-milligram fresh plant organs. Mol. Plant 9, 175–177 (2016).
Hu, J. et al. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20, 320–336 (2008).
Chen, M. et al. Seed fatty acid reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ. 35, 2155–2169 (2012).
Silverstone, A. L. et al. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13, 1555–1566 (2001).
Dill, A., Jung, H. S. & Sun, T. P. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl Acad. Sci. USA 98, 14162–141627 (2001).
Huang, M., Hu, Y., Liu, X., Li, Y. & Hou, X. Arabidopsis LEAFY COTYLEDON1 mediates postembryonic development via interacting with PHYTOCHROME-INTERACTING FACTOR4. Plant Cell 27, 3099–3111 (2015).
Sugliani, M., Rajjou, L., Clerkx, E. J., Koornneef, M. & Soppe, W. J. Natural modifiers of seed longevity in the Arabidopsis mutants abscisic acid insensitive3-5 (abi3-5) and leafy cotyledon1-3 (lec1-3). New Phytol. 184, 898–908 (2009).
White, C. N., Proebsting, W. M., Hedden, P. & Rivin, C. J. Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol. 122, 1081–1088 (2000).
Nadeau, C. D. et al. Tissue-specific regulation of gibberellin biosynthesis in developing pea seeds. Plant Physiol. 156, 897–912 (2011).
Gomez, M. D., Ventimilla, D., Sacristan, R. & Perez-Amador, M. A. Gibberellins regulate ovule integument development by interfering with the transcription factor ATS. Plant Physiol. 172, 2403–2415 (2016).
West, M. A. L. et al. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6, 1731–1745 (1994).
Le, B. H. et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl Acad. Sci. USA 107, 8063–8070 (2010).
Nambara, E., Naito, S. & McCourt, P. A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 2, 435–441 (1992).
Lim, S. et al. ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878 (2013).
Smit, M. E. & Weijers, D. The role of auxin signaling in early embryo pattern formation. Curr. Opin. Plant Biol. 28, 99–105 (2015).
Lee, L. Y. et al. STUNTED mediates the control of cell proliferation by GA in Arabidopsis. Development 139, 1568–1576 (2012).
Hou, X. et al. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5, 4601 (2014).
Zuo, J., Niu, Q.-W. & Chua, N.-H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273 (2000).
Berleth, T. & Jurgens, G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118, 575–587 (1993).
Liu, X. et al. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 7, 12768 (2016).
Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008).
Bailey, T. L. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–169 (2011).
Sauer, M. & Friml, J. In vitro culture of Arabidopsis embryos within their ovules. Plant J. 40, 835–843 (2004).
Acknowledgements
We thank T.-P. Sun for providing ga3ox1/4 and ga3ox1/3/4 seeds and Y. Zhao for pYUC4:GUS seeds. This work was supported by grants from the National Natural Science Foundation of China (31670319) and the Natural Science Foundation of Guangdong Province (2017A030313211).
Author information
Authors and Affiliations
Contributions
Y.H. and X.L.H. conceived and designed the project. Y.H., L.Z., M.H. and Y.Y. conducted the experiments. Y.H., X.L.H., X.L., Y.L. and X.M.H. analysed the data. Y.H. and X.L.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–15 and Supplementary Table 1.
Supplementary Table 2
List of DELLA–LEC1 coregulated genes identified by RNA-seq.
Supplementary Table 3
LEC1 binding peaks and associated genes list of ChIP-seq and LEC1–DELLA targeted genes list.
Supplementary Table 4
List of primers used in this study.
Rights and permissions
About this article
Cite this article
Hu, Y., Zhou, L., Huang, M. et al. Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nature Plants 4, 289–298 (2018). https://doi.org/10.1038/s41477-018-0143-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0143-8
This article is cited by
-
Genome-wide identification, characterization and expression profile analysis of BBX gene family in Chinese chestnut (Castanea mollissima)
Plant Biotechnology Reports (2024)
-
Photoperiod controls plant seed size in a CONSTANS-dependent manner
Nature Plants (2023)
-
Insights from multi-omics integration into seed germination of Taxus chinensis var mairei
Communications Biology (2023)
-
Anatomical and hormonal factors determining the development of haploid and zygotic embryos of oat (Avena sativa L.)
Scientific Reports (2022)
-
Design, Synthesis and Gibberellin-Like Activity of Novel 1-Substituted 3-[3-(Trifluoromethyl)phenyl]thiourea Derivatives
Journal of Plant Growth Regulation (2022)