Abstract
During thymic negative selection, autoreactive thymocytes carrying T cell receptor (TCR) with overtly strong affinity to self-MHC/self-peptide are removed by Bim-dependent apoptosis, but how Bim is specifically regulated to link TCR activation and apoptosis induction is unclear. Here we identify a murine T cell-specific genomic enhancer EBAB (Bub1-Acoxl-Bim), whose deletion leads to accumulation of thymocytes expressing high affinity TCRs. Consistently, EBAB knockout mice have defective negative selection and fail to delete autoreactive thymocytes in various settings, with this defect accompanied by reduced Bim expression and apoptosis induction. By contrast, EBAB is dispensable for maintaining peripheral T cell homeostasis via Bim-dependent pathways. Our data thus implicate EBAB as an important, developmental stage-specific regulator of Bim expression and apoptosis induction to enforce thymic negative selection and suppress autoimmunity. Our study unravels a part of genomic enhancer codes that underlie complex and context-dependent gene regulation in TCR signaling.
Similar content being viewed by others
Introduction
T cell population in the thymus is highly heterogeneous, harboring a diverse T cell receptor (TCR) repertoire1,2,3,4. The massive diversity in TCR sequences is on one hand useful, as it puts the immune system on stand-by for numerous foreign antigens such as pathogens, while on the other hand it is risky because it could generate T cells harboring TCRs that strongly recognize self, potentially causing autoimmunity. To suppress autoimmunity, organisms have evolved a sophisticated mechanism called negative selection, establishing central T cell tolerance. In negative selection, interaction between TCR and self-peptide presented on major histocompatibility complexes (self-pMHCs) is converted into apoptotic output: high-affinity TCR clones are considered as autoreactive and die by apoptosis1,2,3,4.
A part of high-affinity TCR clones, instead of being deleted, are diverted into regulatory T (Treg) cells that are potent to suppress autoreactive T cells in periphery1,5. Suppression by Treg cells is one of the peripheral tolerance mechanisms for organisms to deal with autoreactive T cells that have evaded negative selection. Other mechanisms are induction of T cell anergy and peripheral deletion by apoptosis1.
Pro-apoptotic Bim promotes the mitochondrial apoptosis cascade, contributing to numerous biological pathways6,7,8,9,10,11,12,13,14,15,16,17,18. In central T cell tolerance, Bim is considered as a downstream target of TCR signal: TCR signal activates Bim expression, and Bim knockout (KO) mice show defective negative selection6. However, little is known about how TCR signal strength is linked to Bim expression2,17.
Bim is genetically required not only for establishing central T cell tolerance6,7,8, but also for depleting activated T cells in periphery11,12, B cell homeostasis, embryonic development, and so on18. Therefore, Bim should be able to distinguish multiple biological pathways in different cell types, depending on signals that cells receive. The molecular mechanism underlying how Bim is regulated to work at an appropriate place and time remains elusive.
Enhancers are genomic elements that regulate gene expression in a signal and cell type dependent manner19,20. Although epigenome analyses have enabled systematic identification and characterization of enhancers, it is still difficult to directly study their physiological roles in vivo for the following reasons. First, enhancers are located often several hundreds of kilobases to even megabases away from their target genes, making it difficult to confidently predict a target(s) of an enhancer. Second, some genes may have multiple functionally redundant enhancers. Third, making enhancer KOs through genetic ablation has been labor-intensive and time-consuming, especially in mice. Recent progress in CRISPR–Cas9 technology21 has reduced the cost and time needed for generating enhancer KO mice, and most importantly, has enabled us to produce large genomic deletions without leaving unwanted footprints of exogenous DNAs. CRISPR–Cas9 technology is indeed beginning to uncover physiological functions of novel enhancers in vivo22,23,24.
Here, we utilize enhancer genetics to understand how Bim is specifically regulated to induce apoptosis during thymic negative selection, and find a cis-regulatory enhancer specifically contributing to this process. With the aid of epigenome analyses, we identify a cis-regulatory enhancer EBAB (Bub1-Acoxl-Bim) that is specific to thymocytes and splenic T cells. We generate EBAB KO mice by CRISPR–Cas9 technology and find that a high-affinity TCR repertoire accumulates in the EBAB KO thymus. EBAB KO thymocytes are defective in apoptosis due to incomplete activation of Bim. By contrast, Bim-mediated homeostasis of Treg cells and peripheral T cells is not affected by EBAB KO, thereby implicating a specific function of EBAB in thymic negative selection. This study is an example of utilizing enhancer KO approach to dissect regulation of enhancer activity and subsequent gene function in vivo to address biological questions.
Results
Identification of a murine T cell-specific enhancer E BAB
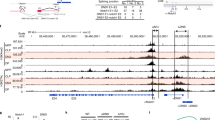
Our analyses on various publicly available ChIP-seq (chromatin-immunoprecipitation and following sequencing) data on multiple mouse tissues identified a T cell-specific enhancer-like region (H3K27ac high and H3K4me3 low)19 in the mouse Bub1-Acoxl-Bim locus (Fig. 1a and Supplementary Fig. 1). This region was located at approximately 200-kb upstream of Bim, within the ninth intron of Acoxl gene (unexpressed in T cells), and at approximately 90-kb upstream of Bub1 (a mitotic checkpoint factor), and thus was named EBAB. EBAB was approximately 8-kb in length and contained two prominent H3K27ac peaks E1 and E2 (Fig. 1a). Both E1 and E2 were highly specific to the thymus (Fig. 1a) and well conserved between human and mice (Fig. 1a and Supplementary Fig. 1b). H3K27ac peaks corresponding to EBAB were identified also in the spleen to a lesser extent (Fig. 1a). The signals in the spleen were likely derived from splenic peripheral T cells because naïve peripheral T cells, but not CD19+ B cells, retained DNase hypersensitivity sites in the locus (Fig. 1b), the observation further supported by other publicly available ChIP-seq data sets (Supplementary Fig. 1c).
Identification of a T cell-specific cis-regulatory element EBAB. a ChIP-seq visualization of H3K27ac in several mouse tissues. H3K27ac profiles from the thymus, spleen, bone marrow, liver, kidney, heart, brain, testis, and brown adipose tissues (BAT) are visualized using the UCSC genome browser (mm9). The EBAB region is highlighted. b DNase hypersensitivity sites (DHS) in the same locus shown in (a). DHS profiles from the thymus, T-Naïve CD4+, regulatory T (Treg) cells, spleen, and B cells (CD43− or CD19+) are visualized using the UCSC genome browser (mm9). The EBAB region is highlighted. c Schematic representation of ΔEBAB. Arrowheads indicate the primers listed in Supplementary Data 1. d Genomic PCR against the EBAB locus of WT, heterozygotes (EBAB+/−), and ΔEBAB mice. A representative gel-image of founder #44-derived DNAs is shown. See also Supplementary Fig. 2 for the results from #47- and #50-derived DNAs. e, f qPCR analysis for Bim (e) and Bub1 (f) on thymocytes, splenocytes, lung, liver, kidney, and pancreas. Data are pooled from five independent experiments (thymocyte, splenocyte; n = 5 WT and EBAB+/−–ΔEBAB littermate pairs, 7–17 weeks old, mean ± s.d.) or three independent experiments (lung, liver, kidney, pancreas; n = 3 WT & EBAB+/−–ΔEBAB littermate pairs, 10–17 weeks old). Each symbol represents an individual mouse; small horizontal lines indicate the mean. No statistically significant differences between WT and EBAB+/− and ΔEBAB were detected (P ≥ 0.05; unpaired two-tailed Student’s t test)
To investigate a physiological role of EBAB in vivo, we generated EBAB KO mice by using the CRISPR–Cas9 system (Fig. 1c and Supplementary Data 1). Three founder lines (line#44, #47 and #50) were successfully obtained, each harboring a distinct pattern of deletion (Fig. 1d and Supplementary Fig. 2). Because offspring from these different founders showed no phenotypic differences (Supplementary Fig. 3), we refer to these three KO alleles simply as ΔEBAB in this paper.
To ask if EBAB KO affects expression of Bim and Bub1, we performed quantitative polymerase chain reaction (qPCR) experiments on several organs and cell types including thymocytes and splenocytes (Fig. 1e, f). Bim and Bub1 expression were slightly decreased in thymocytes and splenocytes in ΔEBAB mice (7–17 weeks old), while unaltered in the lung, liver, kidney, and pancreas (Fig. 1e, f). Thus, EBAB is an enhancer specific to thymocytes and splenic T cells (Fig. 1a, b and Supplementary Fig. 1), deletion of which only slightly affected its proximal genes Bim and Bub1 in total thymocytes and splenocytes (Fig. 1e, f).
E BAB KO accumulates high-affinity TCR clones in the thymus
Next, we asked whether ΔEBAB impairs T cell homeostasis in the thymus. For this purpose, thymocytes were analyzed using flow cytometry with anti-CD4 and anti-CD8 antibodies (Fig. 2a–d). While the extent of reduction in Bim and Bub1 expression was very modest in ΔEBAB thymocytes (Fig. 1e, f), severe abnormalities in thymocyte population were observed both in young (7–17 weeks old) and aged (30–36 weeks old) mice (Fig. 2a, b): ΔEBAB increased proportion of double negative (DN), CD4 single positive (CD4 SP), and CD8 SP thymocytes, whereas proportion of double positive (DP) had decreased (Fig. 2a, b). These were attributed to the increased number of DN, CD4 SP, and CD8 SP thymocytes (Fig. 2c, d). No obvious phenotypic differences were observed between WT and heterozygotes, between ΔEBAB males and ΔEBAB females, and among the three founder lines (Supplementary Fig. 3).
The ΔEBAB thymus accumulates high affinity TCR clones. a Flow cytometric analysis of CD4 versus CD8 thymocyte populations. Data are representative of seven independent experiments (Young; n = 7 WT and EBAB+/−–ΔEBAB littermate pairs, 7–17 weeks old) or five independent experiments (Aged; n = 5 WT & EBAB+/−–ΔEBAB littermate pairs, 30–36 weeks old). b–d Double negative (DN), double positive (DP), CD4 single positive (SP), and CD8 SP thymocyte proportions of young and aged mice (b), and cell numbers of young (c) and aged (d) mice. Data are pooled from seven independent experiments (Young; n = 7 WT and EBAB+/−–ΔEBAB littermate pairs, 7–17 weeks old) or five independent experiments (Aged; n = 5 WT and EBAB+/-–ΔEBAB littermate pairs, 30–36 weeks old). e Scatter plot (log2 fold change versus –log10 P value) of genes analyzed by RNA-seq (n = 2 WT and EBAB+/−–ΔEBAB littermate pairs, 10–11 weeks old). Genes showing more than twofold changes with P < 0.05 (unpaired two-tailed Student’s t test) are highlighted. f Flow cytometric analysis of TCRβ versus CD69 thymocyte populations. Data are representative of five independent experiments (Young; n = 5 WT and EBAB+/−–ΔEBAB littermate pairs, 9–12 weeks old, Aged; n = 6 WT and EBAB+/− mice and n = 7 ΔEBAB mice from WT and EBAB+/−–ΔEBAB littermate pairs and trios, 30–36 weeks old). g Post-selection (TCRβhighCD69high) thymocyte proportion of young and aged mice. Data are pooled from five independent experiments (Young; n = 5 WT and EBAB+/−–ΔEBAB littermate pairs, 9–12 weeks old, Aged; n = 6 WT and EBAB+/− mice, n = 7 ΔEBAB mice, WT and EBAB+/−–ΔEBAB littermate pair or trio, 30–36 weeks old). h qPCR analysis for Nr4a1 in pre-selection (TCRβlowCD69low) and post-selection thymocytes of WT and ΔEBAB. Data are pooled from five independent experiments (n = 5 sex-matched WT–ΔEBAB pairs, 10–18 weeks old, mean ± s.d.). Edges of the box are the 25th and 75th percentiles, and error bars extend to the maximum and minimum. Each symbol in (b, c, d, g) represents an individual mouse; small horizontal lines indicate the mean. n.s. not significant (P ≥ 0.05); *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired two-tailed Student’s t test)
To understand the nature of altered T cell homeostasis in the ΔEBAB thymus, we performed whole transcriptome analysis of ΔEBAB thymocytes (two littermate pairs) utilizing RNA-seq (Fig. 2e). As shown in Fig. 2e, only a small number of genes showed statistically significant (P < 0.05) more than twofold changes (see methods and Supplementary Data 2 for the detail). Yet, differentially expressed genes (DEGs) affected by ΔEBAB indicated that the ΔEBAB thymus accumulated high affinity TCR clones (Fig. 2e): the ΔEBAB thymus exhibited a higher level of Nr4a1 expression when compared to the WT thymus. Nr4a1 is a faithful responder for TCR signal, and the expression level of Nr4a1 positively correlates with TCR signal strength15,25,26. Elevated expression of Nr4a1 is thus one of the hallmarks of high affinity TCR clones in the thymus. Arhgap20 and Tnfrsf9, the top two most elevated genes in RNA-seq data in the ΔEBAB thymus (Fig. 2e), are also known as being upregulated in TCR-activated T cells26, supporting the notion that high-affinity TCR clones accumulate in ΔEBAB mice.
To obtain more evidence that the ΔEBAB thymus piles up high-affinity TCR clones, we stained thymocytes with anti-TCRβ and anti-CD69 antibodies (Fig. 2f, g). We found that proportion of post-selection (TCRβhighCD69high) thymocytes was increased in the ΔEBAB thymus both in young and aged mice (Fig. 2f, g). Moreover, as determined by cell-sorting followed by qPCR, Nr4a1 expression in post-selection thymocytes was much higher in ΔEBAB than in WT (Fig. 2h). Thus, it was likely that high affinity TCR clones were accumulated in the ΔEBAB thymus.
We also analyzed TCR+ population in the DN subset: thymic precursors of TCRαβ+CD8αα+ intestinal intraepithelial lymphocytes (IELs)27,28 and TCR Vα14+ invariant natural killer T cells (iNKT cells)29,30. Our data demonstrated that ΔEBAB increased the number of IEL precursors (Supplementary Fig. 4a, b), while iNKT population was much less affected by ΔEBAB (Supplementary Fig. 4c–e). These changes may contribute to the increased number of DN thymocytes in ΔEBAB mice (Fig. 2c, d).
E BAB is required for eliminating high-affinity TCR clones
Accumulation of high-affinity TCR clones in the ΔEBAB thymus led us to hypothesize that EBAB is required for negative selection and/or more general apoptosis processes. To test this hypothesis, we treated primary thymocytes with four apoptotic stimuli: dexamethasone (DEX)7,8, phorbol 12-myristate 13-acetate (PMA)7,8, ionomycin7,8 (Supplementary Fig. 5), and anti-CD3 and anti-CD28 antibodies6,25 (Fig. 3a). ΔEBAB did not affect survival of DEX- or PMA-treated thymocytes (Supplementary Fig. 5a, b) but partially rescued thymocytes from cell death caused by ionomycin or anti-CD3 and anti-CD28 antibodies (Fig. 3a and Supplementary Fig. 5c–e). The rescue in ionomycin experiments was observed in a dose-dependent manner (Supplementary Fig. 5c–e). PMA, ionomycin, and anti-CD3 and anti-CD28 antibodies are often used for artificially activating TCR stimulation and thus for mimicking negative selection ex vivo6,7,8,25. Thus, these results suggest that ΔEBAB thymocytes were defective in apoptosis following TCR signal activation, leading to a hypothesis that EBAB plays a role in depleting high-affinity TCR clones.
EBAB contributes to depletion of high affinity TCR clones in ex vivo TCR stimulation and the HY tg models. a Annexin V+Propidium Iodide (PI)− fraction of total thymocytes stimulated with anti-CD3 and anti-CD28 antibodies (10 µg/ml) for 9 h. The percentage in mock sample (i.e., background) is subtracted. Data are pooled from 11 independent experiments (n = 3 WT–ΔEBAB littermate pairs, n = 3 sex-matched WT–ΔEBAB pairs, n = 1 WT–Bim−/− littermate pair, n = 1 sex-matched WT–Bim−/− pair, n = 3 sex-matched WT–Bim−/−–ΔEBAB trios, 5–20 weeks old). Edges of the box are the 25th and 75th percentiles, and error bars extend to the maximum and minimum. Outliers are defined as the data point that is located outside of q3 + 1.5(q3 − q1) and q1 − 1.5(q3 − q1), in which q1 and q3 are the 25th and 75th percentiles. b CD4 versus CD8 flow cytometric analysis of PI−HY-TCR+ thymocytes from female and male HY tg mice. The number in the plot is representative percentage of each gate. c−d DP thymocyte proportion (c) and cell numbers (d) of PI−HY-TCR+ thymocytes from female and male HY tg mice. e, f CD8 SP thymocyte proportion (e) and cell numbers (f) of PI−HY-TCR+ thymocytes from female and male HY tg mice. Data are representative of (b), or pooled from (c–f), six independent experiments (n = 1 Bim+/−; HY+ and Bim−/−; HY+ female mice, n = 5 Bim+/−; HY+ and Bim−/−; HY+ male mice, n = 3 EBAB+/−; HY+ and ΔEBAB; HY+ female mice, n = 5 EBAB+/−; HY+ and ΔEBAB; HY+ male mice, 6–8 weeks old). Each symbol in (c–f) represents an individual mouse; small horizontal lines indicate the means. n.s. not significant (P ≥ 0.05); *P < 0.05, **P < 0.01 (unpaired one-tailed Student’s t test or Mann–Whitney U test for Bim KO data, and unpaired two-tailed Student’s t test for ΔEBAB data)
To assess the possibility that EBAB is required for deleting high affinity TCR clones in vivo, we took advantage of three TCR transgenic mouse models: HY transgenic (tg)6 (Fig. 3b–f), OT-II tg6,31,32 (Fig. 4), and OT-I tg33,34,35 (Supplementary Fig. 6).
EBAB is essential for depleting high affinity TCR clones in the OT-II tg model. a CD4 versus CD8 flow cytometric analysis of TCR Vβ5+ thymocytes from mice injected with OVA257–264 or OVA323–229 peptide intraperitoneally. The number in the plot is representative percentage of each gate. b CD4 SP thymocyte proportion of TCR Vβ5+ thymocytes from mice treated as in Fig. 4a. c TCRβ versus CD69 flow cytometric analysis of TCR Vβ5+ thymocytes from mice treated as in Fig. 4a. The number in the plot is representative percentage of the gate. d Post-selection (TCRβhighCD69high) thymocyte proportion of TCR Vβ5+ thymocytes from mice treated as in Fig. 4a. Data are representative of (a, c) or pooled from (b, d) 13 independent experiments (n = 5 WT and EBAB+/− mice injected with OVA257–264 peptide, n = 8 WT and EBAB+/−; OT-II+ mice injected with OVA323–339 peptide, n = 6 ΔEBAB; OT-II+ mice injected with OVA257–264 and OVA323–339 peptide, n = 3 Bim+/−; OT-II+ and Bim−/−; OT-II+ mice injected with each peptide, 5–14 weeks old). Each symbol (b, d) represents an individual mouse; small horizontal lines indicate the mean. n.s. not significant (P ≥ 0.05); **P < 0.01, ***P < 0.001 (unpaired one-tailed Student’s t test for Bim KO data, unpaired two-tailed Student’s t test for ΔEBAB data)
HY tg mice express αβTCRs that recognize a male-specific endogenous antigen called HY, which is presented by H-2Db class I MHC molecules6 (Fig. 3b-f). ΔEBAB; HY tg mice were analyzed at 6–8 weeks old as described previously6 by flow cytometry. As shown by the lower abundance of DP and CD8 SP in EBAB+/−; HY tg male mice compared to female mice (Fig. 3b–f), thymocytes expressing HY-TCR are negatively selected in a male-specific manner. Notably, ΔEBAB significantly rescued DP (both in proportion and number) and CD8 SP (in proportion) thymocytes in HY tg male mice (Fig. 3b–f).
OT-II tg thymocytes express Vα2/Vβ5 TCRs that primarily recognize chicken ovalbumin-derived peptide (ISQAVHAAHAEINEAGR, OVA323–339 peptide) presented by I-Ab class II MHC molecules6,31,32. ΔEBAB; OT-II tg mice were generated by crossing and injected with OVA323–339 peptide or the control peptide intraperitoneally. WT; OT-II tg or EBAB+/−; OT-II tg mice were also used as a control. In response to injection with OVA323–339 peptide, the CD4 SP proportion of control thymocytes was massively reduced (Fig. 4a, b). Strikingly, EBAB KO rescued this reduction (Fig. 4a, b). Staining thymocytes with anti-TCRβ and anti-CD69 antibodies revealed that ΔEBAB almost completely prevented the deletion of post-selection thymocytes caused by OVA323–339 peptide injection (Fig. 4c, d).
Fetal thymic organ culture (FTOC) with the OT-I tg system allows us to investigate effects of antigens of interests on intrathymic T cell development33,34,35,36. OT-I tg thymocytes express Vα2/Vβ5 TCRs that bind chicken ovalbumin peptide residues 257–264 (SIINFEKL, OVA257–264 peptide) in the context of H-2Kb class I MHC molecules34. Of note, a series of OVA257–264 variants with different TCR affinity can be used in the OT-I tg FTOC system34,35. We cultured fetal thymus (FT) of EBAB+/−; OT-I tg and ΔEBAB; OT-I tg in the presence of OVA257–264, Q4R7 (SIIQFERL), or gp33 (KAVYNFATC)34 (Supplementary Fig. 6). The previous publications establish OVA257–264 and Q4R7 as negative selectors (affinity to OT-I TCR: OVA257–264 > Q4R7)34,35. gp33, which does not bind to OT-I TCRs, was used as a control. ΔEBAB compared to EBAB+/− significantly rescued DP thymocytes from Q4R7-dependent selection but not those from OVA257–264-dependent selection (Supplementary Fig. 6). Thus, ΔEBAB rescued OT-I TCR+ thymocytes from negative selection in a TCR affinity-dependent manner.
Taken the results from the three transgenic models together, we concluded that EBAB plays an important role in apoptosis of high-affinity TCR clones in thymic negative selection ex vivo and in vivo.
E BAB is essential for TCR-dependent activation of Bim
The above-described phenotypes observed in the ΔEBAB thymus, four ex vivo culture systems, and three transgenic models are strikingly similar to those observed in Bim KO mice as described in multiple publications6,7,8,9,10 and as validated by ourselves (Figs. 2–4 and Supplementary Figs. 4–7). For example, the extent of increase for post-selection thymocytes caused by ΔEBAB was similar to that by Bim KO (Fig. 2g. versus Supplementary Fig. 7e). Such similarity was also observed for the ex vivo TCR stimulation (Fig. 3a), OT-II (Fig. 4), and OT-I (Supplementary Fig. 6) experiments, whereas Bim KO seemingly more efficiently rescued HY-TCR+ thymocytes than EBAB KO did in the HY experiments (Fig. 3: see discussion regarding interpretation on this data). Moreover, ΔEBAB did not affect DEX-induced cell death, which was in fact Bim-independent (Supplementary Fig. 5a). Some of the thymocytes abnormalities are seen also in mice ectopically expressing Bim-antagonist Bcl2 (Bcl2 tg) in a T cell-specific manner, which leads to defective apoptosis in the thymus25,37,38. Although ΔEBAB had a minor impact on Bim expression in total thymocytes (Fig. 1e), we next examined whether the deficient negative selection of ΔEBAB thymocytes is due to abnormal regulation of Bim. To this end, we again artificially activated TCR by treating thymocytes with anti-CD3 and anti-CD28 antibodies ex vivo, and then measured expression of Bim, Nr4a1, Bub1, and Bcl2 by qPCR (Fig. 5a–d). Bim expression in ΔEBAB thymocytes treated with anti-CD3 and anti-CD28 antibodies was clearly lower than that in TCR-stimulated WT thymocytes (Fig. 5a). Expression of Nr4a1, a faithful responder for TCR signal, was comparable between WT and ΔEBAB (Fig. 5b), suggesting that TCR signal was properly activated in ΔEBAB thymocytes. Consistent with the report that a dominant negative form of Bub1 is insufficient to cause T cell abnormalities39, Bub1 expression was not influenced by TCR stimulation and EBAB KO (Fig. 5c). Bcl2, a TCR-downstream anti-apoptotic gene, exhibited an expected response to TCR stimulation40, and this response was comparable between WT and ΔEBAB (Fig. 5d). Thus, it was likely that EBAB primarily targets Bim following TCR signal.
EBAB regulates Bim expression upon TCR stimulation. a–d Gene expression changes for Bim (a), Nr4a1 (b), Bub1 (c), and Bcl2 (d) in total thymocytes stimulated for 3 h with anti-CD3 and anti-CD28 antibodies (10 µg/ml). e, f Gene expression level (e) and relative gene expression to WT (f) for Bim in pre-selection (TCRβlowCD69low) and post-selection (TCRβhighCD69high) thymocytes. g, h Gene expression level (g) and relative gene expression to WT (h) for Bub1 in pre-selection (TCRβlowCD69low) and post-selection (TCRβhighCD69high) thymocytes. Data are pooled from six independent experiments (a–d; n = 6 sex-matched WT–ΔEBAB pairs, 9–18 weeks old) or five independent experiments (e–h; n = 5 sex-matched WT–ΔEBAB pairs, 10–18 weeks old). Edges of the box are the 25th and 75th percentiles, and error bars extend to the maximum and minimum. Outliers are defined as the data point that is located outside of q3 + 1.5(q3 − q1) and q1 − 1.5(q3 − q1), in which q1 and q3 are the 25th and 75th percentiles. n.s. not significant (P ≥ 0.05); *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired two-tailed Student’s t test)
To assess the role of EBAB in TCR-dependent Bim activation in vivo, we sorted pre- and post-selection thymocytes and measured expression of Bim and Bub1 (Fig. 5e–h). If EBAB regulates Bim in a TCR signal dependent manner, Bim expression should be affected by ΔEBAB at the post-selection stage while less affected at the pre-selection stage. As expected, expression of Bim in the post-selection (TCRβhighCD69high) thymocytes was markedly lower in ΔEBAB than in WT (ΔEBAB/WT = 0.31) (Fig. 5e, f). Although Bim expression was moderately affected by EBAB KO in the pre-selection (TCRβlowCD69low) stage (ΔEBAB/WT = 0.64) (Fig. 5e, f), expression ratio of ΔEBAB to WT in the post-selection stage was significantly lower than that in the pre-selection stage (Fig. 5f). These results were validated at the protein level, demonstrating that protein expression of BimEL and BimL, two of major isoforms of Bim41, was compromised by ΔEBAB (Supplementary Fig. 9). Together, ΔEBAB affected Bim expression in a post-selection-biased manner, indicating an important role of EBAB in activating Bim upon TCR activation in vivo. Bub1 expression was not significantly altered between ΔEBAB and WT mice both in pre- and post-selection thymocytes (Fig. 5g, h), again excluding Bub1 in explaining the T cell phenotypes we observed (Figs. 2–4). Collectively, our data demonstrated that EBAB KO disrupts transcriptional activation for Bim upon TCR stimulation, resulting in the rescue of high affinity TCR clones in the thymus (Figs. 2–5). We reasoned that Bim expression in total thymocytes looked only mildly affected by EBAB KO (Fig. 1e) as only 10–30% of thymocytes is at the post-selection stage where transcriptional regulation of Bim is strongly affected by ΔEBAB.
Furthermore, we investigated whether EBAB regulates Bim in cis or trans. To test this, we generated EBAB+/−; Bim−/+ mice (Supplementary Fig. 10a). If EBAB controls Bim in cis, a phenotype of EBAB+/−; Bim−/+ thymocytes should be similar to ΔEBAB (Supplementary Fig. 10a). If EBAB can regulate Bim in trans, EBAB+/−; Bim−/+ mice should show no phenotype in the thymus (Supplementary Fig. 10a). Staining with anti-CD4, anti-CD8, anti-TCRβ, and anti-CD69 antibodies revealed that EBAB+/−; Bim−/+ thymocytes exhibited ΔEBAB-like phenotypes (Supplementary Fig. 10b–f). Given that Bim+/− exhibits almost no phenotype in DN, DP, CD4 SP, CD8 SP, and TCRβhighCD69high thymocytes (Supplementary Fig. 10g), these results validated that EBAB is a cis-regulatory element regulating Bim.
E BAB is dispensable for Treg cells and peripheral T cells
Expression levels of Nr4a transcription factors including Nr4a1 are markers not only for high affinity TCR clones, but also for thymic Treg cells that suppress autoreactive T cells in periphery1,5. Nr4a transcription factors cooperatively promote Treg differentiation through directly activating expression of Foxp3, the master regulator for Treg identity42,43,44. Thus, it is thought that high-affinity TCR clones are a precursor of Treg cells5. Bim KO and Bcl2 tg mice accumulate both of high affinity TCR clones and Treg cells6,7,8,14,15,25,37,38 (Supplementary Figs. 7d, e, 8c). To examine whether accumulation of high-affinity TCR clones resulted in the increased number of Treg cells in ΔEBAB mice, we analyzed thymic and splenic Treg cells by staining with anti-Foxp3 antibody (Fig. 6a, b). We found that the number of thymic and splenic Treg cells were comparable between WT and ΔEBAB (Fig. 6a, b). Thus, although high-affinity TCR clones are accumulated in the ΔEBAB thymus (Fig. 2), EBAB does not affect homeostasis of Treg cells. These results indicate that Bim-dependent homeostasis of Treg cells in the thymus and spleen is independent on EBAB.
EBAB does not play a major role in maintaining homeostasis of regulatory T cells and peripheral T cells. a, b CD4+Foxp3+ thymic (a) and splenic (b) Treg cell numbers of young and aged mice. Data are pooled from five independent experiments (Young; n = 5 WT and EBAB+/−–ΔEBAB littermate pairs, 9–12 weeks old, Aged; n = 5 WT EBAB+/−–ΔEBAB littermate pairs, 30–36 weeks old). c Flow cytometric analysis of CD4 versus CD8 T cell populations in the spleen. The number in the plot is representative percentage of each gate. Data are representative of five independent experiments (Young; n = 5 WT and EBAB+/−–ΔEBAB littermate pairs, 7–17 weeks old, Aged; n = 5 WT and EBAB+/−–ΔEBAB littermate pairs, 30–36 weeks old). d CD4+ and CD8+ splenic T cell numbers. Data are pooled from five independent experiments (Young; n = 5 WT and EBAB+/−–ΔEBAB littermate pairs, 7–17 weeks old, Aged; n = 5 WT and EBAB+/−–ΔEBAB littermate pairs, 30–36 weeks old). Each symbol in (a, b, d) represents an individual mouse; small horizontal lines indicate the mean. n.s. not significant (P ≥ 0.05) (unpaired two-tailed Student’s t test)
Bim KO and Bcl2 tg mice also accumulate peripheral T cells6,7,8,12,13,25,37,38 (Supplementary Fig. 8a, b). However, the number of splenic CD4+ and CD8+ T cell were not affected by ΔEBAB both in young and aged mice (Fig. 6c, d). In addition, splenic B cells, whose homeostasis depends on Bim8, were unaffected by EBAB KO (Supplementary Fig. 11). These results indicated that homeostasis of peripheral T cells is properly maintained in ΔEBAB mice in contrast to Bim KO mice6,7,8,12,13 (Supplementary Fig. 8a, b).
To gain an additional insight into a role of EBAB in peripheral T cell homeostasis, we performed Interleukin-2 (IL-2) deprivation experiments and activation-induced cell death (AICD) experiments in cultured activated CD4+ and CD8+ splenocytes (Fig. 7).
EBAB is dispensable for two apoptotic pathways in peripheral T cells. a, b Viability (% Annexin V−PI−) of activated CD4+ (a) and CD8+ (b) splenic T cells after IL-2 withdrawal. Data are pooled from ten independent experiments (a; n = 5 sex-matched WT–ΔEBAB pairs and n = 5 sex-matched WT–Bim−/−–ΔEBAB trios, 5–26 weeks old, mean ± s.d.) or 11 independent experiments (b; n = 5 sex-matched WT–ΔEBAB pairs and n = 5 sex-matched WT–Bim−/−–ΔEBAB trios, 5–26 weeks old, mean ± s.d.). c, d Gene expression level for Bim in activated CD4+ (c) and CD8+ (d) splenic T cells cultured with or without IL-2 for 6 h. Data are pooled from five independent experiments (n = 5 sex-matched WT–Bim−/−–ΔEBAB trios, 5–14 weeks old). e, f Expression of Bub1 in activated CD4+ (e) and CD8+ (f) splenic T cells cultured with or without IL-2 for 6 h. Data are pooled from five independent experiments (n = 5 sex-matched WT–Bim−/−–ΔEBAB trios, 5–14 weeks old). g, h Apoptotic cell rate (%Annexin V+) of activated CD4+ (g) and CD8+ (h) splenic T cells restimulated with anti-CD3 antibody (5 µg/ml) for 6 h. Data are pooled from five independent experiments (n = 5 sex-matched WT–Bim−/−–ΔEBAB trios, 5–26 weeks old). Edges of the box are the 25th and 75th percentiles, and error bars extend to the maximum and minimum. Outliers are defined as the data point that is located outside of q3 + 1.5(q3 − q1) and q1 − 1.5(q3 − q1), in which q1 and q3 are the 25th and 75th percentiles (c–h). n.s. not significant (P ≥ 0.05); *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired two-tailed Student’s t test)
Bim-deficient peripheral T cells are resistant to the absence of IL-2 ex vivo7,45 while WT and ΔEBAB splenic T cells needed IL-2 for their continuous survival in culture (Fig. 7a, b). In accordance with this, EBAB was dispensable for upregulation of Bim following IL-2 withdrawal (Fig. 7c, d). Expression of Bub1 was merely affected by IL-2 (Fig. 7e, f). Hence, IL-2 deprivation-dependent T cell death in periphery requires Bim but not EBAB.
AICD is known as a peripheral cell death cascade where repeatedly activated T cells undergo apoptosis46. In fact, reactivating peripheral T cells by anti-CD3 antibody treatment strongly induced cell death (Fig. 7g, h). Using this system, we found that neither Bim nor EBAB was required for AICD ex vivo (Fig. 7g, h).
From these experiments, we concluded that at least two peripheral apoptotic pathways are intact in ΔEBAB mice, which can be one explanation for why the increase in thymic high-affinity TCR clones did not lead to accumulation of peripheral T cells.
We additionally analyzed splenic activated T cell proportion by staining splenocytes with anti-CD44 and anti-CD62L antibodies to investigate autoimmune pathology (Fig. 8a–c). In line with the fact that two peripheral apoptosis pathways are intact in ΔEBAB mice (Fig. 7), activated T cell (CD44highCD62Llow) proportion was not increased in the ΔEBAB spleen even in aged mice (Fig. 8a–c and Supplementary Fig. 12a–e). These data were in line with RNA-seq data from WT and ΔEBAB spleen (two littermate pairs), where only two candidate DEGs were identified (Supplementary Fig. 12f and Supplementary Data 3). In contrast, Bim-deficient mice accumulated activated CD8+ T cells in the spleen already at younger age (Supplementary Fig. 8d–f).
ΔEBAB mice do not show any autoimmune phenotype. a Flow cytometric analysis of CD62L versus CD44 populations in CD4+ or CD8+ splenic T cell. The number in the plot is representative percentage of each gate. For CD4+ T cells, gates indicate activated (CD44highCD62Llow) and naïve (CD44lowCD62Lhigh) populations. For CD8+ T cells, activated effector memory (CD44highCD62Llow), activated central memory (CD44highCD62Lhigh) and naïve (CD44lowCD62Lhigh) populations are gated. Data are representative of four independent experiments (n = 5 for WT and EBAB+/− mice, n = 6 for ΔEBAB mice from WT & EBAB+/−–ΔEBAB littermate pairs and trios, 30–34 weeks old). b, c Activated CD4+ (b) and CD8+ (c) proportions of aged mice. Data are pooled from four independent experiments (n = 5 for WT and EBAB+/− mice, n = 6 for ΔEBAB mice from WT and EBAB+/−–ΔEBAB littermate pairs and trios, 30–34 weeks old). d The mean (±s.e.m.) clinical scores at the days after EAE was induced in WT (control) (n = 10) and ΔEBAB mice (n = 8). The incidence of EAE: control 10/10, ΔEBAB 8/8. No data point showed statistically significant difference between WT and ΔEBAB (i.e., unpaired two-tailed Student’s t test P ≥ 0.05). e Representative pictures for Hematoxylin and Eosin staining for the lung, liver, kidney, and pancreas of two independent experiments (n = 2 WT and EBAB+/−–ΔEBAB littermate pairs, 30–31 weeks old). The scale bars represent 100 μm. Each symbol in (b, c) represents an individual mouse; small horizontal lines indicate the mean. n.s. not significant (P ≥ 0.05) (unpaired two-tailed Student’s t test)
Furthermore, we assessed a role of EBAB in an in vivo autoimmune disease model, experimental autoimmune encephalomyelitis (EAE)47. As shown in Fig. 8d, clinical scores of ΔEBAB were comparable to those of WT: ΔEBAB mice neither exhibited severer nor ameliorated EAE phenotypes, suggesting that EBAB is dispensable for EAE. In contrast, we confirmed previous publication that Bim plays a role in EAE (Supplementary Fig. 13)16. Consistently, we did not find any histological sign for autoimmunity such as massive infiltration of leukocytes (i.e., inflammation) in several nonlymphoid organs in ΔEBAB mice (Fig. 8e).
Thus, ΔEBAB mice did not show any autoimmune phenotype, which was consistent with that EBAB was not essential for peripheral T cell homeostasis (Figs. 6–8, Supplementary Fig. 8, and Supplementary Figs. 12 and 13). These results highlighted an extraordinary specialized role of EBAB in thymic negative selection (Fig. 9).
Discussion
It is thought that TCR signal activates Bim to promote apoptosis in thymic negative selection process1,2,3. However, it has been unclear how TCR signal strength is linked to the Bim expression level. Even whether activation of Bim expression is critical for negative selection has been unanswered due to lack of a mutant specifically deficient for TCR-dependent Bim expression17. Here, by means of CRISPR–Cas9-mediated KO approach in mice, we generated a mutant that lacks a cis-regulatory enhancer for Bim, EBAB (Figs. 1–5 and Supplementary Fig. 10). EBAB is T cell-specific and evolutionarily conserved (Fig. 1a, b and Supplementary Fig. 1), and ΔEBAB mice were unable to efficiently eliminate high affinity TCR clones (Figs. 2–4 and Supplementary Fig. 6). This defect appeared to be attributed to insufficient activation of Bim during TCR activation (Fig. 5 and Supplementary Fig. 9). Thus, we propose that EBAB links TCR signal and Bim expression, contributing to depletion of high affinity TCR clones in the thymus and thus to central T cell tolerance (Fig. 9).
Bim KO affects homeostasis of nearly all of T cell types6,7,8,9,10,11,12,13,14,15,16,17,18 (Supplementary Fig. 4 and Supplementary Figs. 7 and 8). Comparing T cell phenotypes between Bim KO and ΔEBAB mice provides insights into the extent of contribution of EBAB-dependent Bim regulation on phenotypes of interests. Among the phenotypes observed in the ΔEBAB thymus, accumulation of post-selection thymocytes in the ΔEBAB thymus was as severe as that in the Bim KO thymus (Fig. 2f, g and Supplementary Fig. 7d, e). Moreover, both of ΔEBAB and Bim KO rescued thymocytes from negative selection in the ex vivo TCR stimulation model and three transgenic systems (Figs. 3 and 4 and Supplementary Fig. 6). The degrees of rescues by EBAB KO and Bim KO were similar in the ex vivo TCR stimulation, OT-I, and OT-II experiments (Figs. 3 and 4 and Supplementary Fig. 6). In contrast, in the HY experiments, it appeared that Bim KO better rescued HY-TCR+ thymocytes than EBAB KO. When interpreting these results, it should be noted that readouts used (e.g., proportion of cell types of interests) do not always specifically reflect negative selection. In particular, Bim KO is able to rescue apoptosis induced by various stimuli18, even death of control-treated cells (Supplementary Fig. 5). Thus, increased survival seen in Bim KO potentially reflects both negative selection-dependent and -independent events such as post-selection lifetime. Importantly, our analyses showed that function of EBAB appears to be specific for TCR signal-dependent Bim expression in the thymus. Hence, we assume that the more efficient rescue observed in Bim KO in the HY experiments may be attributed to pleiotropic effects of Bim KO on negative selection-independent phenomena in the thymus. Alternatively, the observations in the HY experiments imply that there may be another enhancer controlling TCR-dependent expression of Bim. Collectively, we suggest that a role for Bim in eliminating high affinity TCR clones in the thymus requires EBAB. These results establish EBAB-mediated transcriptional regulation for Bim as a contributor for Bim-dependent thymic negative selection.
Bim KO and Bcl2 tg mice accumulate Treg cells, a basis for the current model that Treg cells are differentiated from high-affinity TCR clones in the thymus1,5,15,25 (Supplementary 8c). Intriguingly, in the ΔEBAB thymus, the number of Treg cells was comparable with that in the littermate controls even though high affinity TCR clones were increased (Fig. 2f, g and Fig. 6a, b). This indicates that the residual amount of Bim in the ΔEBAB thymus is sufficient for maintaining Treg homeostasis. It is also plausible that a non-cell autonomous mechanism may be dominant for Treg homeostasis in the thymus. Supporting this, a previous study shows that Foxp3 overexpression induces Bim phosphorylation and enhances apoptosis in newly arising thymic Treg cells48. In addition, Treg homeostasis is controlled by availability of pro-survival cytokines such as IL-2 derived from the surrounding environment48,49. These altogether indicate that Treg apoptosis is precisely tuned by multiple mechanisms independent from EBAB, and demonstrate that the role of EBAB is specific for thymic negative selection (Fig. 9). Alternatively, the increased proportion of IEL precursors both in Bim−/− and ΔEBAB mice (Supplementary Fig. 4a, b) suggests that EBAB controls fate determination of survived high-affinity TCR clones: ΔEBAB may bias fate of thymocytes escaped from negative selection toward IEL rather than Treg cells.
Bim KO and Bcl2 tg mice show abnormalities not only in thymic negative selection, but also in peripheral T cell homeostasis6,7,8,11,12,13,25,37,38 (Supplementary Fig. 8a, b and Supplementary Fig. 12). In contrast, peripheral T cells did not accumulate in the ΔEBAB spleen (Fig. 6c, d). Consistent with this, ΔEBAB affects neither peripheral T cell activation nor experimental autoimmunity (Fig. 8 and Supplementary Fig. 12). This suggests that high-affinity TCR clones were peripherally depleted in ΔEBAB mice. One possible explanation for this observation is that peripherally activated T cells properly undergo IL-2 withdrawal-dependent apoptosis, a process that requires Bim but not EBAB (Fig. 7a, b). This idea is further supported by that Bim was properly upregulated upon IL-2 withdrawal in activated T cells even in the absence of EBAB (Fig. 7c, d). Additional peripheral T cell-specific enhancer(s) for Bim, or completely different mechanisms such as post-translational modifications, may also ensure Bim-mediated maintenance for peripheral T cell homeostasis. A different apoptosis pathway such as the Fas pathway could also be a major contributor in eliminating peripheral autoreactive T cells1,46,50,51. The Fas pathway is known to be required for AICD, which we found requires neither Bim nor EBAB (Fig. 7g, h). These results are consistent with that deletion of EBAB had a very minor impact on genome-wide gene expression in peripheral T cells (Supplementary Fig. 12f). Collectively, our findings emphasize that the role of EBAB is dedicated to thymic negative selection (Fig. 9).
A physiological contribution of a gene in a certain biological phenomenon depends on quantity of functional gene products that exist in that context. Regarding classical protein-coding genes, concentration of functional gene products-(modified) proteins is affected by transcriptional regulation, mRNA stability, translational efficacy, post-translational modifications, and protein stability, which are interconnected with each other. Each of these mechanisms profoundly contributes to determine the steady-state concentration of gene products, depending on genes and biological contexts. However, it has been technically difficult to dissect the relative importance of each mechanism for a gene to exert its physiological role(s) in a certain biological situation. Our study is unique in that we disrupted the single gene expression program by targeting the specific cis-element. This enabled us to reveal the degrees of contribution of Bim and EBAB in thymic negative selection measured by different readouts, which represent cutting-edge molecular dissection of the cis-regulatory control for thymic negative selection in vivo. Although we do not exclude a possibility that downstream mechanisms such as Bim phosphorylation might be also important, we provide the evidence that the enhancer-driven gene expression regulation of Bim is an important upstream mechanism in thymic negative selection. In addition, the present work provides an example for using enhancer KO approach, instead of conventional genetic-manipulation of protein-coding genes, to address important biological questions.
Methods
Mice
All animal protocols were approved by the Animal Care and Use committee of Advanced Telecommunications Research Institute International (permission numbers: AN20140002, AN20150002, AN20160002, AN20170002, and AN20180002) and Kyoto university (permission numbers: K-16-12-11 and L-18-1). For generating ΔEBAB mice, four gRNAs were individually cloned into the pX330 vector (#42230, Addgene, MA, USA) that harbors a Cas9 transgene. Briefly, two pairs of oligos (Supplementary Data 1) were annealed at 95 °C followed by natural cooling to 25 °C, and ligated with BbsI (New England Biolabs, MA, USA)-digested pX330 by using Ligation high (TOYOBO, Osaka, Japan). The obtained plasmids were sequence-validated (FASMAC, Kanagawa, Japan). The four gRNAs were together injected into fertilized eggs of C57BL/6N mice at the concentration of 2.5 μg/μl each (Transgenic, Fukuoka, Japan), generating the three different founders #44, #47, and #50. These founders were crossed with WT to obtain F1. Deleted regions were validated by sequencing analyses (FASMAC) against DNA from F0 and F1. The obtained F1 offspring were then crossed to generate ΔEBAB mice. Bim−/− (B6.129S1-Bcl2l11tm1.1Ast/J) and OT-II tg (B6.Cg-Tg(TcrαTcrβ)425Cbn/J) mice were purchased from The Jackson Laboratory (ME, USA). OT-II tg, OT-I tg and HY tg mice were crossed with ΔEBAB and Bim−/− mice for negative selection assays.
In all mice experiments, KO and the littermate control (WT or heterozygote), thus in total two to three mice were sacrificed and analyzed side-by-side on the same day to be considered as a littermate pair or trio. All experiments were performed without randomization. Blinding was performed in the injection process of OT-II negative selection assay. In other experiments, blinding was not done.
DNA isolation and genomic PCR
Genomic DNAs were prepared by using MightyAMP DNA polymerase kit (TaKaRa, Shiga, Japan) according to the manufacturer’s instruction. Genomic PCR experiments were performed using MightyAMP DNA polymerase or KOD FX-neo (TOYOBO) and the primers listed in Supplementary Data 1.
Epigeome analysis
Epigenome analyses were performed with the UCSC genome browser (https://genome.ucsc.edu/). For analyzing epigenome from peripheral naïve T cells, ChIP-seq data retrieved (accession numbers: GSE67443 and GSE60005) were mapped to the mm9 of mouse genome by using Bowtie252. The obtained sam format files were converted into bam format files by using samtools53 that were then subjected to peak calling with MACS54. Wig format files were then uploaded into the UCSC genome browser to be visualized.
RNA isolation, cDNA synthesis, and qPCR
For nonlymphoid organs, total RNAs were prepared by using Trizol reagent (Thermo Fisher Scientific, MA, USA). The obtained supernatants containing total RNAs were further purified with RNeasy mini kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s instruction. Total RNAs from lymphocyte samples were prepared by RNeasy mini kit with the equipped lysis buffer. 0.1–2 μg of total RNAs were then reverse transcribed with SuperScript III first-strand synthesis system (Thermo Fisher Scientific) according to the manufacturer’s instruction. The obtained cDNAs were 5- or tenfold-diluted and subjected into qPCR experiments by using LightCycler480 Instrument II system and SYBR Green Master Mix (Roche, Basel, Switzerland). The obtained data were analyzed using the delta-Ct method.
Antibodies
Anti-CD4 antibody (Clone: GK1.5 (FITC), SONY, Tokyo, Japan), anti-CD8a antibody (Clone: 53-6.7 (APC), SONY), anti-CD8b antibody (eBioH35-17.2 (PE), Invitrogen, CA, USA), anti-TCRβ antibody (Clone: H57-597 (FITC), SONY), anti-TCRβ antibody (Clone: H57-597 (APC/Cy7), BioLegend, CA, USA), anti-CD69 antibody (Clone: H1.2F3 (APC), BioLegend), anti-CD5 antibody (Clone: 53-7.3 (APC), BioLegend), anti-CD44 antibody (Clone: IM7 (Alexa Fluor 700), BioLegend), anti-CD62L antibody (Clone: MEL-14 (PE), BioLegend), anti-TCR Vβ5.1, 5.2 antibody (Clone: MR9-4 (PE), BioLegend), anti-TCR HY antibody (Clone: T3.70 (PE), eBioscience, CA, USA), anti-B220 antibody (Clone: RA3-6B2 (FITC), SONY), LEAF-purified anti-CD3ε antibody (Clone: 145-2C11, BioLegend), LEAF-purified anti-CD28 antibody (Clone: 37.51, BioLegend), Biotin anti-TCRβ antibody (Clone: H57-597, BioLegend) and Biotin anti-CD69 antibody (Clone: H1.2F3, BioLegend) were used in this study.
Flow cytometry
The thymus and spleen were harvested to obtain single lymphocytes suspension. For splenocytes, red blood cells (RBCs) were lysed with RBC lysis buffer (0.015 M NH4Cl, 0.1 mM KHCO3, 0.01 mM Na2EDTA). Live cell numbers were counted by using Countess system (Thermo Fisher Scientific). The obtained samples (1 × 106 cells) were then stained with anti-CD4 antibody (1:200), anti-CD8a antibody (1:100), anti-CD8b antibody (1:200), anti-TCRβ antibody (1:200 (FITC), 1:100 (APC/Cy7)), anti-CD69 antibody (1:100), anti-CD5 antibody (1:100), anti-CD44 antibody (1:200), anti-CD62L antibody (1:100), anti-TCR Vβ5.1, 5.2 antibody (1:100), anti-TCR HY antibody (1:200) or anti-B220 antibody (1:100) in 100 μl of FCM buffer (10% fetal bovine serum (FBS) in phosphate-buffered saline (PBS)) for >30 min at 4 °C in the dark. For IEL precursors and iNKT measurement, 1 × 106 thymocytes were stained with α-GalCer-loaded CD1d tetramer (TS-MCD-1 (PE), MBL, Aichi, Japan) in 50 μl of FCM buffer for >30 min at 4 °C in the dark, and then stained with additional antibodies (see Supplementary Fig. 4 for the details). To analyze Treg cells, the obtained single cell suspension (3–5 × 106 cells) was stained using Foxp3 staining kit (Clone: FJK-16s, eBioscience, CA, USA), essentially according to the manufacturer’s instruction. Cell fixation was performed for 10 min. Samples were analyzed on EC800 (SONY) and FACS CantoII (BD Bioscience, NJ, USA). Gating strategies are shown in Supplementary Fig. 14.
RNA-seq and bioinformatic analysis
Total RNAs were extracted from the thymus and spleen as described in the RNA isolation section. RNA-seq libraries were generated using the SureSelect Strand-Specific RNA Library Prep for Illumina (Agilent) according to the manufacturer’s instructions. Sequencing experiments were performed with Hiseq2500 (Illumina; Single End 36 bp). The obtained reads were mapped to the mouse genome mm9 using Illumina Eland with the default parameter setting. Uniquely aligned reads were retrieved allowing up to 2 bp mismatches, and the number of exon-mapped overlapping reads were counted. The obtained gene list with reads per million per a kilobase (RPKM) scores were shown in Supplementary Datas 2 and 3. To identify DEGs, we first focused on the well-annotated protein-coding genes. RPKM scores from two replicates were averaged, and the ratio ΔEBAB/WT were calculated. In this calculation, 1 was added to all averaged RPKM scores to ignore scores below “1”, and to make analyses more stringent. The obtained ratios were used to sort genes to find candidate DEGs, followed by statistical analyses (unpaired two-tailed Student’s t-test) and qPCR validation.
Cell sorting
To purify pre- and post-selection thymocytes, single cell suspension (1–2 × 107 cells) was stained with anti-TCRβ antibody (1:100 or 1:200) and anti-CD69 antibody (1:100 or 1:200) in 100 μl of FCM buffer for >30 min at 4 °C in the dark. The stained cells were then tenfold diluted and sorted using SH800 cell sorter system (SONY).
Ex vivo thymocytes cell death assay
Totally, 1 × 105 total thymocytes were cultured with DEX (10 nM), PMA (2 ng/ml) or ionomycin (1 μg/ml or 0.1 μg/ml) in 200 μl of RPMI1640 media (10% FBS, 1% penicillin streptomycin, 50 μM 2-mercaptoethanol, 1 × nonessential amino acids (nacalai tesque, Kyoto, Japan) and 1 × sodium pyruvate (nacalai tesque)) in a 96-well flat bottom plate. DMSO- or EtOH-treated cells served as controls. Viability was measured by Annexin V FLUOS staining kit (Roche) at days 0–2.
Ex vivo TCR stimulation
Ex vivo TCR stimulation experiments were performed essentially as described previously7,25. Briefly, 12-well plates were coated with LEAF-purified anti-CD3ε and anti-CD28 antibodies (0 or 10 μg/ml in 500 μl of PBS, BioLegend) for 2 h at 37 °C. After 3 times-wash with 0.5 ml of PBS, 1 × 106 cells of total thymocytes were cultured in 2 ml of DMEM media (10% FBS, 1% penicillin streptomycin, 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 10 mM HEPES (pH 7.4), 1 × nonessential amino acids (nacalai tesque) and 1 × sodium pyruvate (nacalai tesque)) for 3 h (for qPCR experiments) or 9 h (for Annexin V and PI staining using Annexin V FLUOS staining kit (Roche)).
Peptides
OVA323–339 peptide (ISQAVHAAHAEINEAGR, BEX, Tokyo, Japan), OVA257–264 peptide (SIINFEKL, BEX), gp33 (KAVYNFATC, BEX), and Q4R7 (SIIQFERL, BEX) were certified as >98% pure by HPLC. Peptides were dissolved in dimethyl sulfoxide (DMSO) at concentration of 100 mg/ml and stored at −80 °C. The peptide/DMSO solutions were diluted in PBS just before the experiment.
OT-II negative selection assay
OT-II negative selection assay was performed as described previously6. Briefly, WT; OT-II+, EBAB+/−; OT-II+ and ΔEBAB; OT-II+ mice were injected intraperitoneally with 1 mg OVA323–339 peptide or OVA257–264 peptide as a control resuspended in 500 μl of PBS. Thymocytes were harvested after 72 h for analysis.
OT-I tg fetal thymic organ culture
FTOC was performed as described previously33,34,35. Briefly, fetal thymic lobes were excised at embryonic day 15 (E15) and cultured on Whatman Nuclepore Track-Etched Membrane (WHA110409, GE Healthcare, Little Chalfont, England) floated on RPMI1640 media (10% FBS, 1% penicillin streptomycin, 50 μM 2-mercaptoethanol, 1 × nonessential amino acids (nacalai tesque, Kyoto, Japan) and 1 × sodium pyruvate (nacalai tesque)) in the presence of 2 μM OVA257–264, 2 μM Q4R7, or 20 μM gp33. On day 4, thymocytes were analyzed by flow cytometry.
Immunoblotting assay
CD69+ and CD69− T cells were enriched from thymocytes using biotinylated anti-CD69 antibody (BioLegend) and MojoSort Streptavidin Nanobeads (BioLegend). CD69- T cells were further purified by depleting TCRβ+ cells using biotinylated anti-TCRβ antibody (BioLegend) and MojoSort Streptavidin Nanobeads (BioLegend). Expression of Bim and ACTB proteins in each subset was detected with anti-Bim antibody (1:1000, #2819, Cell Signaling, MA, USA) and anti-ACTB antibody (1:10,000, NB600-532, Novus Biologicals, CO, USA). As the secondary antibody, anti-rabbit IgG (1:15,000, 711-035-152, Jackson ImmumoResearch, PA, USA) was used. Signals were visualized with ECL plus Western Blotting Detection Reagents (GE Healthcare) and analyzed by the CCD digital imaging system LAS-4000 Luminescent Image Analyzer (GE Healthcare). Whole proteins were stained with SYPRO Ruby Protein Gel Stain (S12000, Thermo Fisher Scientific) and detected by the CCD digital imaging system LAS-4000 Luminescent Image Analyzer (GE Healthcare). Uncropped scans are available in the Source data file.
Cytokine withdrawal assay
CD4+ or CD8+ T cells were isolated from splenocytes using MojoSort Mouse CD4 or CD8 T Cell Isolation Kit (BioLegend), respectively according to the manufacturer’s instruction. T cells were activated by culturing cells in RPMI1640 media (10% FBS, 1% penicillin streptomycin, 50 μM 2-mercaptoethanol, 1 × nonessential amino acids (nacalai tesque) and 1 × sodium pyruvate (nacalai tesque)) plus IL-2 (100 U/ml, Wako, Osaka, Japan) in 96 well flat bottom plate coated with LEAF-purified anti-CD3ε and anti-CD28 antibodies (5 μg/ml in 50 μl of PBS, BioLegend) for 48 hr. Activated T cells were then cultured for 1 day with RPMI1640 plus IL-2 (100 U/ml, Wako) in normal 96-well plate. After that, IL-2 was removed from the media and viability of CD4+ and CD8+ T cells was measured using Annexin V FLUOS staining kit (Roche) on days 1–3 after IL-2 withdrawal.
Activation induced cell death assay
Isolation and activation of CD4+ or CD8+ T cells were performed as described in the “cytokine withdrawal assay” section. After activation and 1-day culture in the presence of IL-2, T cells were further incubated in RPMI1640 media (10% FBS, 1% penicillin streptomycin, 50 μM 2-mercaptoethanol, 1 × nonessential amino acids (nacalai tesque) and 1 × sodium pyruvate (nacalai tesque)) without IL-2 in a 96-well flat bottom plate coated with LEAF-purified anti-CD3ε (5 μg/ml in 50 μl of PBS, BioLegend) for 6 h. Cell viability was determined using Annexin V FLUOS staining kit (Roche).
Histochemistry
Mouse tissues were fixed in 10% neutral buffered formalin. Samples were then processed into 6 micron sections, mounted, and Hematoxylin and eosin (H&E)-stained (Genostaff, Tokyo, Japan). The prepared samples were observed with Nikon Ni-E microscope (Nikon, Tokyo, Japan).
Experimental autoimmune encephalomyelitis
Experimental autoimmune encephalomyelitis (EAE) experiments were performed essentially as described previously47. Briefly, mice (WT, ΔEBAB, and Bim−/−) were subjected to subcutaneous injection at two different sites with an emulsion of complete Freund’s adjuvant (CFA; 250 μg heat-killed Mycobacterium tuberculosis H37Ra (Difco) dissolved in 50 μl incomplete Freund’s adjuvant (Sigma)) and 250 μg myelin oligodendrocyte glycoprotein (MOG) peptides (amino acids 35–55) in 50 μl PBS. Mice were then injected with 200 ng of Pertussis Toxin (Calbiochem) intraperitoneally at day 0 and day 2 after EAE induction. The clinical scores were assessed as follows: (1) flaccid tail, (2) impaired righting reflex and/or gait, (3) partial hind limb paralysis, (4) total hind limb paralysis, and (5) total hind limb paralysis with partial forelimb paralysis.
Statistics
The sample size was chosen as follows. First, the number of animals was minimized as much as possible in light of animal ethics. Second, against effect size estimated in each experiment, ≥80–90% power was favored. Third, in most cases, n = 5 was set as a threshold according to the previous report55. These three criterion functioned to determine the sample size as n = 5–8 in most experiments. Moreover, dot-plot/box plot representation of data provided insights into how the samples were distributed, and thus into the extent of difference of two groups. This, in the specific cases, led us to conclude that relatively small number of animals (i.e., less than five animals) was enough to support our conclusion.
According to the observed differences, our sample size appeared to be appropriate to meet≥80–90% power. In addition, dot-plot/box plot representation of data was useful to estimate sample distribution. Clearly abnormal distribution was not observed in our experiments, further supporting that our statistical tests were appropriate.
Significant differences between two groups were examined using one or two-tailed, unpaired t test, or Mann–Whitney U test. One-tailed test was chosen when we had hypothesis and/or previous knowledge regarding direction of changes (e.g., increased or decreased) in experiments.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The RNA-seq data have been deposited in DNA Data Bank of Japan (DDBJ) under the accession code DRA004726 and DRA008123. Source data underlying graphs and blots are supplied as a Source data file. All other data are included in the supplemental information or available from the authors upon reasonable requests.
References
Xing, Y. & Hogquist, K. A. T-cell tolerance: central and peripheral. Cold Spring Harb. Perspect. Biol. 4, a006957 (2012).
Klein, L., Kyewski, B., Allen, P. M. & Hogquist, K. A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14, 377–391 (2014).
Hogquist, K. A. & Jameson, S. C. The self-obsession of T cells: how TCR signaling thresholds affect fate ‘decisions’ and effector function. Nat. Immunol. 15, 815–823 (2014).
Richards, D. M., Kyewski, B. & Feuerer, M. Re-examining the nature and function of self-reactive T cells. Trends Immunol. 37, 114–125 (2016).
Wirnsberger, G., Hinterberger, M. & Klein, L. Regulatory T-cell differentiation versus clonal deletion of autoreactive thymocytes. Immunol. Cell Biol. 89, 45–53 (2011).
Bouillet, P. et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 415, 922–926 (2002).
Bouillet, P. et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286, 1735–1738 (1999).
Labi, V. et al. Deregulated cell death and lymphocyte homeostasis cause premature lethality in mice lacking the BH3-only proteins Bim and Bmf. Blood 123, 2652–2662 (2014).
Herold, M. J. et al. Impact of conditional deletion of the pro-apoptotic BCL-2 family member BIM in mice. Cell Death Dis. 5, e1446 (2014).
Li, K. P. et al. Temporal expression of Bim limits the development of agonist-selected thymocytes and skews their TCRbeta repertoire. J. Immunol. 198, 257–269 (2017).
Hildeman, D. A. et al. Activated T cell death in vivo mediated by proapoptotic bcl-2 family member bim. Immunity 16, 759–767 (2002).
Hale, J. S., Nelson, L. T., Simmons, K. B. & Fink, P. J. Bcl-2-interacting mediator of cell death influences autoantigen-driven deletion and TCR revision. J. Immunol. 186, 799–806 (2011).
Li, W. Q., Guszczynski, T., Hixon, J. A. & Durum, S. K. Interleukin-7 regulates Bim proapoptotic activity in peripheral T-cell survival. Mol. Cell Biol. 30, 590–600 (2010).
Chougnet, C. A. et al. A major role for Bim in regulatory T cell homeostasis. J. Immunol. 186, 156–163 (2011).
Stritesky, G. L. et al. Murine thymic selection quantified using a unique method to capture deleted T cells. Proc. Natl Acad. Sci. USA 110, 4679–4684 (2013).
Ludwinski, M. W. et al. Critical roles of Bim in T cell activation and T cell-mediated autoimmune inflammation in mice. J. Clin. Invest. 119, 1706–1713 (2009).
Strasser, A., Puthalakath, H., O’Reilly, L. A. & Bouillet, P. What do we know about the mechanisms of elimination of autoreactive T and B cells and what challenges remain. Immunol. Cell Biol. 86, 57–66 (2008).
Sionov, R. V., Vlahopoulos, S. A. & Granot, Z. Regulation of Bim in health and disease. Oncotarget 6, 23058–23134 (2015).
Calo, E. & Wysocka, J. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837 (2013).
Vernimmen, D. & Bickmore, W. A. The hierarchy of transcriptional activation: from enhancer to promoter. Trends Genet. 31, 696–708 (2015).
Sander, J. D. & Joung, J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355 (2014).
Canver, M. C. et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015).
LaFlam, T. N. et al. Identification of a novel cis-regulatory element essential for immune tolerance. J. Exp. Med. 212, 1993–2002 (2015).
Thomas, G. D. et al. Deleting an Nr4a1 super-enhancer subdomain ablates ly6c(low) monocytes while preserving macrophage gene function. Immunity 45, 975–987 (2016).
Burger M. L., Leung K. K., Bennett M. J., Winoto A. T cell-specific inhibition of multiple apoptotic pathways blocks negative selection and causes autoimmunity. Elife 3, 03468 (2014). https://doi.org/10.7554/eLife.03468
Fassett, M. S., Jiang, W., D’Alise, A. M., Mathis, D. & Benoist, C. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion. Proc. Natl Acad. Sci. USA 109, 3891–3896 (2012).
Konkel, J. E. et al. Control of the development of CD8alphaalpha+ intestinal intraepithelial lymphocytes by TGF-beta. Nat. Immunol. 12, 312–319 (2011).
McDonald, B. D., Bunker, J. J., Ishizuka, I. E., Jabri, B. & Bendelac, A. Elevated T cell receptor signaling identifies a thymic precursor to the TCRalphabeta(+)CD4(−)CD8beta(−) intraepithelial lymphocyte lineage. Immunity 41, 219–229 (2014).
Matsuda, J. L. et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med. 192, 741–754 (2000).
Pellicci, D. G. et al. A natural killer T (NKT) cell developmental pathway Involving a thymus-dependent NK1.1(−)CD4(+) CD1d-dependent precursor stage. J. Exp. Med. 195, 835–844 (2002).
Kisielow, P., Bluthmann, H., Staerz, U. D., Steinmetz, M. & von Boehmer, H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+ 8+ thymocytes. Nature 333, 742–746 (1988).
Barnden, M. J., Allison, J., Heath, W. R. & Carbone, F. R. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76, 34–40 (1998).
Hogquist, K. A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994).
Daniels, M. A. et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729 (2006).
Takada, K. et al. TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8(+) T cells. Nat. Immunol. 16, 1069–1076 (2015).
Nitta, T., Ohigashi, I. & Takahama, Y. The development of T lymphocytes in fetal thymus organ culture. Methods Mol. Biol. 946, 85–102 (2013).
Strasser, A., Harris, A. W., von Boehmer, H. & Cory, S. Positive and negative selection of T cells in T-cell receptor transgenic mice expressing a bcl-2 transgene. Proc. Natl Acad. Sci. USA 91, 1376–1380 (1994).
Strasser, A., Harris, A. W. & Cory, S. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell 67, 889–899 (1991).
Cowley, D. O., Muse, G. W. & Van Dyke, T. A dominant interfering Bub1 mutant is insufficient to induce or alter thymic tumorigenesis in vivo, even in a sensitized genetic background. Mol. Cell Biol. 25, 7796–7802 (2005).
Schmitz, I., Clayton, L. K. & Reinherz, E. L. Gene expression analysis of thymocyte selection in vivo. Int Immunol. 15, 1237–1248 (2003).
O’Connor, L. et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17, 384–395 (1998).
Sekiya, T. et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2, 269 (2011).
Sekiya, T. et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 14, 230–237 (2013).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Kelly, P. N. et al. Individual and overlapping roles of BH3-only proteins Bim and Bad in apoptosis of lymphocytes and platelets and in suppression of thymic lymphoma development. Cell Death Differ. 17, 1655–1664 (2010).
Krammer, P. H., Arnold, R. & Lavrik, I. N. Life and death in peripheral T cells. Nat. Rev. Immunol. 7, 532–542 (2007).
Yasuda, K. et al. Satb1 regulates the effector program of encephalitogenic tissue Th17 cells in chronic inflammation. Nat. Commun. 10, 549 (2019).
Tai, X. et al. Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity 38, 1116–1128 (2013).
Lio, C. W. & Hsieh, C. S. A two-step process for thymic regulatory T cell development. Immunity 28, 100–111 (2008).
Fortner, K. A., Bouillet, P., Strasser, A. & Budd, R. C. Apoptosis regulators Fas and Bim synergistically control T-lymphocyte homeostatic proliferation. Eur. J. Immunol. 40, 3043–3053 (2010).
Ju, S. T. et al. Fas(CD95)/FasL interactions required for programmed cell death after T-cell activation. Nature 373, 444–448 (1995).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Krzywinski, M. & Altman, N. Points of significance: comparing samples—part I. Nat. Methods 11, 215–216 (2014).
Acknowledgements
We thank Dr. Thomas. N. Sato (T.N.S), the director of The TNS BioMEC-X Laboratories, ATR, and JST ERATO Sato Live Bio-forecasting project, for supporting all aspects of the study. We thank Tomoko Kuroda, Tomoko Ninomiya, Hitomi Anabuki, Satsuki Endo, Fumihiko Sagawa, Satoshi Kozawa, Terumi Horiuchi, and Kiyomi Imamura for technical assistance. We thank Ryoko Takahashi, Erika Kojima, and Toshiya Morie for administrative assistance. We are thankful to Dr. Pieter Bas Kwak and Dr. Bryce Nelson for critically reading the manuscript. We also thank Dr. Kosuke Yusa and Dr. Takeshi Watanabe for providing insightful comments on the manuscript. Dr. Thomas. N. Sato and the members of the TNS BioMEC-X Laboratories provided insightful comments on the manuscript. This work was supported by JST ERATO (to S.K. via T.N.S; JPMJER1303), JSPS KAKENHI (26890030, 15H01478, 18K15409, and 18H04810: S.K.), The Uehara Memorial Foundation (S.K.), The Shimizu Foundation for Immunology and Neuroscience Grant for 2018 (S.K) and Japan Foundation for Applied Enzymology (S.K.).
Author information
Authors and Affiliations
Contributions
M.A.H. performed and analyzed the most of the experiments except EAE and wrote the paper. K.M., Y.N. and H.K. performed the TCR transgenic mice experiments. H.H., K.Y., D.O., Y.T. and K.H. performed the EAE experiments. Y.S. and S.K. performed the RNA-seq and bioinformatics analyses. S.K. conceived and supervised the project, and wrote the paper. All authors provided intellectual input and reviewed the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information: Nature Communications thanks Stephen Jameson and other anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source Data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hojo, M.A., Masuda, K., Hojo, H. et al. Identification of a genomic enhancer that enforces proper apoptosis induction in thymic negative selection. Nat Commun 10, 2603 (2019). https://doi.org/10.1038/s41467-019-10525-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-019-10525-1
This article is cited by
-
Remote solid cancers rewire hepatic nitrogen metabolism via host nicotinamide-N-methyltransferase
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.