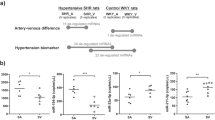
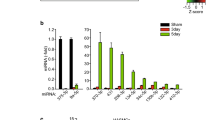
Abstract
Endothelial cells, which regulate arterial stiffness via endothelial-derived substances, are independently and strongly associated with hypertension. However, the exact roles of exosome miRNAs from brain endothelial cells in the development of hypertension are still not fully explored. Here, we investigated the miRNA functions systematically by examining both exosomal small RNA and mRNA of endothelial cells in Wistar Kyoto (WKY) rats versus spontaneously hypertensive rats (SHRs). Our findings revealed that miRNAs, representing ~60–70%, account for the majority of small RNAs. Moreover, we found 159 novel miRNAs in total from the unannotated reads across the diverse samples. Afterwards, 76 differentially expressed miRNAs (37 upregulated, 39 downregulated) and 1709 differentially expressed mRNAs (775 upregulated, 934 downregulated) were identified between SHRs and WKY rats, respectively. Finally, 647 genes targeted by 36 miRNAs came to our attention via identification of the target genes of those abnormal miRNAs. The differentially expressed target genes induced by miRNA changes were mapped to a number of genes involved in various gene functions and pathways. These changes lead to dysregulation of angiogenesis, axonogenesis, neuron-to-neuron synapses, focal adhesion, axon guidance, cell adhesion molecules (CAMs), adherens junction, and ECM-receptor interaction pathways. Together, our study revealed that the miRNAs are changed and contribute to the dysregulated functions and pathways of their target genes and provided more insights into their regulation mechanisms during mammalian hypertension development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Cohuet G, Struijker-Boudier H. Mechanisms of target organ damage caused by hypertension: therapeutic potential. Pharmcol Ther. 2006;111:81–98.
Serne EH, de Jongh RT, Eringa EC, RG IJ, Stehouwer CD. Microvascular dysfunction: a potential pathophysiological role in the metabolic syndrome. Hypertension. 2007;50:204–11.
Wilkinson IB, Franklin SS, Cockcroft JR. Nitric oxide and the regulation of large artery stiffness: from physiology to pharmacology. Hypertension. 2004;44:112–6.
Targonski PV, Bonetti PO, Pumper GM, Higano ST, Holmes DR Jr., Lerman A. Coronary endothelial dysfunction is associated with an increased risk of cerebrovascular events. Circulation. 2003;107:2805–9.
Yuan X, Wu Q, Liu X, Zhang H, Xiu R. Transcriptomic profile analysis of brain microvascular pericytes in spontaneously hypertensive rats by RNA-Seq. Am J Transl Res. 2018;10:2372–86.
Pietri P, Vyssoulis G, Vlachopoulos C, Zervoudaki A, Gialernios T, Aznaouridis K, et al. Relationship between low-grade inflammation and arterial stiffness in patients with essential hypertension. J Hypertens. 2006;24:2231–8.
Recio-Mayoral A, Rimoldi OE, Camici PG, Kaski JC. Inflammation and microvascular dysfunction in cardiac syndrome X patients without conventional risk factors for coronary artery disease. JACC Cardiovasc Imaging. 2013;6:660–7.
Booth AD, Wallace S, McEniery CM, Yasmin, Brown J, Jayne DR, et al. Inflammation and arterial stiffness in systemic vasculitis: a model of vascular inflammation. Arthritis Rheum. 2004;50:581–8.
Fichtlscherer S, Rosenberger G, Walter DH, Breuer S, Dimmeler S, Zeiher AM. Elevated C-reactive protein levels and impaired endothelial vasoreactivity in patients with coronary artery disease. Circulation. 2000;102:1000–6.
Greenberg JI, Shields DJ, Barillas SG, Acevedo LM, Murphy E, Huang J, et al. A role for VEGF as a negative regulator of pericyte function and vessel maturation. Nature. 2008;456:809–13.
Stapor PC, Sweat RS, Dashti DC, Betancourt AM, Murfee WL. Pericyte dynamics during angiogenesis: new insights from new identities. J Vasc Res. 2014;51:163–74.
Garcia-Contreras M, Shah SH, Tamayo A, Robbins PD, Golberg RB, Mendez AJ, et al. Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration Type 1 diabetes. Sci Rep. 2017;7:5998.
Ebrahimkhani S, Vafaee F, Young PE, Hur SSJ, Hawke S, Devenney E, et al. Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci Rep. 2017;7:14293.
Alipoor SD, Tabarsi P, Varahram M, Movassaghi M, Dizaji MK, Folkerts G, et al. Serum exosomal miRNAs are associated with active pulmonary tuberculosis. Dis Markers. 2019;2019:1907426.
Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112:532–8.
Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH, et al. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci Rep. 2015;5:11295.
Bhome R, Del Vecchio F, Lee GH, Bullock MD, Primrose JN, Sayan AE, et al. Exosomal microRNAs (exomiRs): small molecules with a big role in cancer. Cancer Lett. 2018;420:228–35.
van Balkom BW, de Jong OG, Smits M, Brummelman J, den Ouden K, de Bree PM, et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. 2013;121:3997–4006. S3991-3915.
Bovy N, Blomme B, Freres P, Dederen S, Nivelles O, Lion M, et al. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget. 2015;6:10253–66.
Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24.
Nakagawa S, Deli MA, Nakao S, Honda M, Hayashi K, Nakaoke R, et al. Pericytes from brain microvessels strengthen the barrier integrity in primary cultures of rat brain endothelial cells. Cell Mol Neurobiol. 2007;27:687–94.
Wu Q, Jing Y, Yuan X, Li B, Wang B, Liu M, et al. The distinct abilities of tube-formation and migration between brain and spinal cord microvascular pericytes in rats. Clin Hemorheol Microcirc. 2015;60:231–40.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–41.
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8.
Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1.
Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
The Gene Ontology C. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–8.
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–61.
Spijkers LJ, van den Akker RF, Janssen BJ, Debets JJ, De Mey JG, Stroes ES, et al. Hypertension is associated with marked alterations in sphingolipid biology: a potential role for ceramide. PLoS ONE. 2011;6:e21817.
Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90.
Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–21.
Liu X, Yuan W, Yang L, Li J, Cai J. miRNA profiling of exosomes from spontaneous hypertensive rats using next-generation sequencing. J Cardiovasc Transl Res. 2019;12:75–83.
Santovito D, Mandolini C, Marcantonio P, De Nardis V, Bucci M, Paganelli C, et al. Overexpression of microRNA-145 in atherosclerotic plaques from hypertensive patients. Expert Opin Ther Targets. 2013;17:217–23.
Zagrean AM, Hermann DM, Opris I, Zagrean L, Popa-Wagner A. Multicellular crosstalk between exosomes and the neurovascular unit after cerebral ischemia. Therapeutic implications. Front Neurosci. 2018;12:811.
Acknowledgements
This study was supported by the innovation fund of the Chinese Academy of Medical Sciences and Peking Union Medical College (Nos. 3332014006 and 3332015123), the CAMS Initiative for Innovative Medicine (CAMS-I2M) (No. 2016-I2M-3-006), and the National Natural Science Foundation of China (81801433).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wu, Q., Yuan, X., Li, B. et al. Integrated exosomal miRNA and transcriptome analysis of brain microvascular endothelial cells in spontaneously hypertensive rats. Hypertens Res 43, 90–98 (2020). https://doi.org/10.1038/s41440-019-0345-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41440-019-0345-0
Keywords
This article is cited by
-
Circulating exosomal MicroRNAs: New non-invasive biomarkers of non-communicable disease
Molecular Biology Reports (2021)