Abstract
The conserved microRNA396 (miR396) is involved in plant growth, development, and abiotic stress response in multiple plant species through regulating its targets, Growth Regulating Factor (GRF) transcription factor genes. However, the role of miR396 has not yet been characterized in perennial monocot species. In addition, the molecular mechanism of miR396-mediated abiotic stress response remains unclear. To elucidate the role of miR396 in perennial monocot species, we generated transgenic creeping bentgrass (Agrostis stolonifera) overexpressing Osa-miR396c, a rice miRNA396 gene. Transgenic plants exhibited altered development, including less shoot and root biomass, shorter internodes, smaller leaf area, fewer leaf veins, and epidermis cells per unit area than those of WT controls. In addition, transgenics showed enhanced salt tolerance associated with improved water retention, increased chlorophyll content, cell membrane integrity, and Na+ exclusion during high salinity exposure. Four potential targets of miR396 were identified in creeping bentgrass and up-regulated in response to salt stress. RNA-seq analysis indicates that miR396-mediated salt stress tolerance requires the coordination of stress-related functional proteins (antioxidant enzymes and Na+/H+ antiporter) and regulatory proteins (transcription factors and protein kinases). This study establishes a miR396-associated molecular pathway to connect the upstream regulatory and downstream functional elements, and provides insight into the miRNA-mediated regulatory networks.
Similar content being viewed by others
Introduction
Soil salinity is a major constraint for crop productivity. High Na+ content during salt stress results in ionic stress due to the disturbed intracellular ion homeostasi1. Excess Na+ negatively affects K+ uptake because of their similar chemical properties, and thereby leads to the inhibition of many K+-dependent biological processes, such as protein synthesis, enzymatic reactions, and photosynthesis. To alleviate ionic stress and maintain a high K+:Na+ ratio, plants have evolved adaptive strategies of cytosolic Na+ exclusion and vacuolar Na+ sequestration2. Currently, several classes of Na+ transporters have been identified to play central roles in these adaptive strategies during high salinity exposure3,4. The best-characterized Na+ transporters, which alleviate excess Na+ through compartmentalizing Na+ into the vacuoles are NHX transporters via mediating intracellular Na+/H+ and K+/H+ antiport5. Studies in various plant species show that overexpression of NHX genes confers enhanced high salinity tolerance and elevated relative Na+ content in plant tissues6,7. Besides Na+ sequestration, the mechanism of Na+ exclusion has been well elucidated through characterization of salinity overly sensitive 1 (SOS1) transporters, which mediate Na+/H+ antiport across the plasma membrane8. The SOS signaling pathway in response to salt stress has been proposed in Arabidopsis, in which the calcium-binding protein AtSOS3 activates the kinase activity of AtSOS2 to further activate AtSOS1 through the direct phosphorylation9,10. Overexpression of AtSOS1 has been shown to lead to enhanced salt tolerance in transgenic (TG) Arabidopsis11.
Salt stress also induces osmotic stress due to the accumulation of Na+ in the apoplast, thereby elevating osmotic gradient between inside and outside of the cells. In response to osmotic stress, certain organic solutes called osmoprotectants (such as proline, sugars, mannitol, etc.) are produced in the cytoplasm to maintain cell turgor pressure12. A previous study indicated that TG rice accumulating sugar trehalose exhibits enhanced abiotic stress tolerance13.
In addition, salt stress leads to enhanced production of reactive oxygen species (ROS), such as superoxide, hydrogen peroxide, hydroxyl radical, and singlet oxygen, which trigger cell oxidative damage and ultimately cell death14. Antioxidant enzymes, such as superoxide dismutase, catalase, guaiacol peroxidase, ascorbate peroxidase, dehydroascorbate reductase, and glutathione reductase, play an essential role in scavenging the overproduced ROS. Overexpression of genes encoding these antioxidants confers enhanced tolerance to salt and other abiotic stresses in a variety of plant species15,16.
Besides these functional proteins, regulatory roles of transcription factors (TFs), protein kinases, and phosphatases have also been well documented in the plant response to salt stress. Recently, the regulatory role of microRNAs (miRNAs) in plant response to salt and other environmental stresses has been gradually revealed. For example, TG Arabidopsis with higher expression levels of miR408 exhibits enhanced tolerance to salt, cold, and oxidative stresses, but reduced tolerance to drought and osmotic stresses17. MiR408-mediated plant abiotic stress response is associated with enhanced cellular antioxidant capacity and reduced ROS production17. Another recent study shows that constitutive expression of Osa-miR528 confers enhanced salt and nitrogen-deficiency tolerance in TG creeping bentgrass, which is associated with up-regulation of K+ transporter gene HAK5, the increased activities of nitrite reductase and antioxidant enzymes, altered expression of other stress-related TF and small RNAs, and the repression of its direct targets18. These results strongly suggest that miRNAs coordinate multiple stress-responsive pathways to cope with abiotic stress.
MiR396 is a conserved miRNA, and presents in both monocots and dicots. It performs post-transcriptional gene regulation through repressing its targets, Growth Regulating Factors (GRFs). The role of miR396 in plant growth and development has been well characterized. Morphologically, TG plants constitutively expressing miR396 display shorter plants and narrower leaves than WT controls in Arabidopsis and tobacco because of reduced cell numbers in leaf19,20,21. A similar phenotype was observed in atgrf1 atgrf2 atgrf3 triple mutants19. In addition, TG tobacco overexpressing miR396 showed cotyledon fusion and lack of a shoot apical meristem22. A recent study showed that AtGRF5 participates in the control of leaf senescence23. TG Arabidopsis overexpressing miR396 exhibits an early senescence phenotype23. MiR396 is also involved in plant response to various abiotic stresses. At-miR396 was induced under UV-B radiation resulting in inhibited cell division in proliferating tissues24. Expression of tomato miR396a (Sp-miR396) is up-regulated under salt and drought stresses25,26. TG tobacco overexpressing Sp-miR396a exhibits enhanced tolerance to salt, drought, and cold due to the improved osmoregulation and decreased accumulation of ROS26. In contrast, constitutive expression of Osa-miR396c in TG rice and Arabidopsis results in reduced salt and alkali stress tolerance in comparison with wild-type (WT) plants27. The opposite responses to salt stress in different plant species suggest a species-specific function of miR396. Currently, the role of miR396 has not been elucidated in perennial monocot species. In addition, the underlying molecular mechanisms of miR396-mediated plant resistance to salt and other environmental stresses remain unclear.
In this study, we generated TG creeping bentgrass plants constitutively expressing rice miR396 to investigate the role of miR396 in plant development and response to salt stress in this perennial monocot species. MiR396 TG plants display altered leaf morphology and tillering, and improved salt stress tolerance in comparison with WT controls. Furthermore, genome-wide analysis in miR396 TG plants vs. WT controls elucidates the possible regulatory pathway for miR396-mediated salt or other environmental stress responses, which provides insight into the central role of miR396 in the regulatory network.
Materials and methods
Generation of TGs overexpressing MiR396
A 510 bp DNA fragment of Os-miR396c gene (GenBank: AK062523.1) containing pre-miR396c stem-loop structure was amplified from rice cDNA. The cDNAs were then cloned into the pGEM-T Easy vector (Promega, Madison, WI). T Easy-cDNA with the correct sequence was digested and recombined into the binary vector pZH0128 generating an overexpression gene construct, which was then introduced into creeping bentgrass cultivar ‘Penn A-4’ (supplied by HybriGene) via Agrobacterium tumefaciens strain LBA4404-mediated plant transformation. Creeping bentgrass transformation using mature seed-initiated embryonic callus was as described previously29,30.
Plant growth and salt stress treatment
WT and the regenerated TG creeping bentgrass were clonally propagated and grown in cone-tainers (4.0 × 20.3 cm, Dillen Products) filled with pure silicon sand. The plants were fertilized every other day with 0.2 g/L 20:10:20 water-soluble fertilizer (Peat-Lite Special; The Scotts Company) and maintained in a growth room with light regime (14-h of light/ 10-h of dark). Temperatures in the growth room were 25 °C during the light period and 17 °C during the dark period with 350–450 μmol m−2 s−1 light intensity. WT and TG plants were propagated at the same time and from the same amount of tillers to ensure that they were at the same developmental stage for morphological analysis and salt stress treatment.
Salt stress treatment and leaf sample collection for gene expression analysis were performed as described previously with modifications18. In this study, the concentration of NaCl solution was 250 mM. The duration for salt stress treatment was 8 days followed by a 10-day recovery.
Measurement of mineral content, leaf RWC, chlorophyll, and proline content
WT and TG plant leaf tissues were collected before and after 8-day, 250 mM NaCl treatments for the measurement of Na+ and K+ relative contents, leaf RWC, EL, total chlorophyll content, and proline content using previously published protocols31.
Histology analysis
Histological analysis of the leaf and stem cross sections was performed as described previously with modifications18. In this study, sections were stained using Toluidine Blue or Safranin. For leaf epidermis cells observation, clear nail polish was applied to the leaf upper epidermis of the representative WT and TG plants. After drying completely, the films were peeled off and observed under microscope (MEIJI EMZ-5TR).
Plant RNA isolation and expression analysis
Plant total RNA was isolated from 100 mg of the first and second topmost fully expanded leaves of each tiller. RNA isolation, semi-quantitative RT-PCR and stem-loop RT-qPCR analyses were performed according to previously published protocol18. AsUBQ5 (JX570760) was used as an endogenous control. All primers used for sequence amplification, quantitative, and semi-quantitative RT-PCR analysis are listed in supplementary Table 1.
cDNA library preparation and Illumina sequencing, differential expression and GO enrichment analyses
Total RNA of shoots from non-stressed WT and TG plants were extracted and purified using the RNeasy Plant Mini Kit (Qiagen, Germantown, MD). cDNA libraries were constructed using TruSeq RNA Library Preparation v2 (Illumina Inc., San Diego, CA) according to the manufacturer’s protocol. Paired-end sequencing of each library was performed using the HiSeq 2000 (Illumina Technologies) platform following the manufacturer’s instructions (101-bp paired-end reads).
The differential gene expression analysis was performed using R 3.2.0 (https://www.r-project.org) and the Bioconductor package edgeR with FDR-corrected P-value cut-off of <0.05. A MDS plot was generated using edgeR to show the separation between WT and TG samples and consistency between replicates. A volcano plot was created using edgeR to plot log2 FC and the –log10 P-value in TG vs. WT. The heatmaps showing expression profiles between WT and TG samples were generated based on the log2-transformed count values using R’s pHeatmap.2 package (https://cran.r-project.org/web/packages/pheatmap/index.html).
To gain information on the over-represented functional categories, GO-enrichment analysis was performed. Since there is no GO annotation available for creeping bentgrass transcripts, putative GO terms were assigned using NCBI-blast+ and Blast2GO 2.832,33.
Results
MiR396 is regulated by salt stress
Previous studies showed that miR396 responds to salt stress in various plant species, displaying differential expression profiles. For example, miR396 is down-regulated under salt stress in rice, Spartina alternaiflora, Populus cathayana, and Salix matsudanabut Koidz, but up-regulated in tomato26,27,34,35. To investigate the role of miR396 in response to salt stress in a perennial grass species, creeping bentgrass, we set to first study how miR396 responds to salt stress through analyzing its expression profile. Stem-loop RT-qPCR analysis showed that miR396c was significantly up-regulated upon salt treatment (Fig. S1). Although declined 6 h after treatment, it remained noticeably elevated compared to 0 h control (Fig. S1). The result suggests that miR396 might act as a positive regulator in plant salt stress response.
Generation of TG creeping bentgrass constitutively expressing Osa-miR396c
After confirming that miR396c responds to salt stress, we generated TG creeping bentgrass constitutively expressing a rice gene, Osa-miR396c to further study the role of miR396 in plant adaption to salt stress (Fig. S2a). The constitutive expression construct of miR396 was introduced into wild type (WT) plants via A. tumefaciens-mediated transformation. The selectable marker gene, Hyg conferring hygromycin resistance was amplified from the genomic DNA of regenerated TG plants and WT controls for TG event selection (Fig. S2b). The expression levels of pri-miR396c and mature miR396c were then compared between WT and TG plants to determine whether the rice pri-miR396c was successfully integrated into the genome of creeping bentgrass, transcribed, and properly processed (Fig. S2, c and d). We generated a total of 32 individual TG events, which exhibit small leaf area and slow growth rate in comparison with WT controls.
TG creeping bentgrass overexpressing MiR396c exhibits altered plant development
To study if miR396 is implicated in plant development of perennial grass species, 10-week-old WT and TG plants each initiated from a single tiller were compared. As shown in Fig. 1a, TG plants produced smaller shoots and fewer roots than those of WT controls. As a result, TG plants had significantly reduced biomass accumulation in both shoot and root compared to WT controls (Fig. 1f, g). Further analysis of tiller growth indicates that the reduced shoot biomass in TG plants compared to WT controls is associated with shorter tiller length and/or lower tiller number than WT controls (Fig. 1c, h). The average length of the internodes from the longest TG tillers is significantly reduced compared with that of the WT tillers (Fig. 1b, d, and i), resulting in reduced tiller length in TGs. When comparing leaf morphology between WT and TG plants, TG leaves are narrower and shorter than WT leaves (Fig. 1b, e, j, and k), similar to the morphology changes observed in TG Arabidopsis and tobacco constitutively expressing miR39611,21. The result suggests that the role of miR396 in controlling leaf development is conserved between monocots and dicots.
a Ten-week-old WT and TG plants both initiating from a single tiller. Scale bar, 10 cm. b Representative leaves and internodes of WT and TG plants after initiation from single tillers. Scale bar, 2 cm. c Close up of the longest tillers from WT and TG plants. Scale bar, 5 cm. d All internodes from the representative longest tiller were sliced from top to bottom and arranged from left to right. Scale bar, 5 cm. e Three of the topmost fully developed leaves from the representative tillers of WT and TG plants. Scale bar, 2 cm. f Shoot and g root dry weight of WT and TG plants at 10 weeks after initiation from a single tiller (n = 4). h Tiller number in WT and TG plants at 5 and 10 weeks after initiation from a single tiller (n = 5). i Average length of the topmost eight internodes from WT and TG tillers (n = 6). j Leaf length and k leaf width from the representative WT and three transgenic lines (n = 3). Data are shown as means, and error bars represent standard error. A significant difference between WT and each TG line was indicated with asterisks (*, **, or ***) at P < 0.05, 0.01, or 0.001 by Student’s t-test
Previous study indicated that miR396 is involved in cell proliferation21. To study the impact of miR396 on plant development at cellular level, we cross-sectioned leaves and stems in WT and TG plants followed by histological analysis. As shown in Fig. 2a, c, two TG lines have significantly fewer leaf veins than WT controls, resulting in narrower leaves. TG2 plants with FL phenotype displayed reduced stem diameter and a reduced number of vascular bundles, while TG7 plants with the WL phenotype did not show a significant difference from WT controls (Fig. 2b, d, and e). To investigate if the smaller leaf size in TG plants is attributed to the reduced cell proliferation, we compared the number of leaf epidermis cells in WT and TG plants. The result shows that the leaf epidermis cells in TG plants are significantly reduced compared to WT controls (Fig. 2f, g), implying that miR396 is a negative regulator in cell proliferation.
a Cross-section images of WT and TG leaves. Scale bar, 100 µm. b Statistical analysis of leaf vein number between representative WT and TG plants (n = 5). c The representative leaf epidermis of WT and two TG lines. Scale bar, 50 µm. d The number of leaf epidermis cells between WT and two TG lines (n = 5). Data are shown as means with standard error. Asterisks (***) indicate a significant difference between WT and each TG line at P < 0.001 by Student’s t-test
MiR396 TG plants exhibit enhanced salt tolerance associated with improved water retention and cell membrane integrity, increased chlorophyll contents, and decreased proline contents under salt stress
To study the role of miR396 in plant response to salt stress in creeping bentgrass, we chose to use two low transgene expressers, TG7 and TG14 with moderately altered morphology for analysis. Both WT and TG plants initiating from the same number of tillers were mowed to the same height prior to salinity exposure. Plants were then treated by 250 mM NaCl for 8 days followed by a 10-day recovery. As shown in Fig. 5a, upon NaCl treatment and during plant recovery, most of the WT leaves senesced and turned yellow, whereas the TG leaves remained green with little damage (Fig. 3a), indicating that overexpression of miR396c leads to enhanced salt tolerance in TG plants.
a WT and TG plants were subjected to 250 mM NaCl treatment for 8 days followed by a 10-day recovery. b Leaf RWC, c leaf electrolyte leak, d total chlorophyll content, and e proline content were measured in WT and TG leaves at 0 day and 8 days of salt stress treatment. DW dry weight. Data are shown as means (n = 4) with standard error. Asterisks (***) indicate a significant difference between WT and each TG line at P < 0.001 by Student’s t-test
To further investigate what causes the enhanced salt tolerance in TG plants at physiological level, we analyzed leaf relative water content (RWC), leaf electrolyte leakage (EL), total chlorophyll content, and proline content in WT and TG plants before and 8 days after 250 mM NaCl treatment. Before the treatment, WT plants and two TG lines have similar RWC (Fig. 3b). Upon exposure to salt stress, although the RWC of both WT and TG plants declined, the decline in TG plants was significantly less pronounced than that of WT controls (Fig. 3b), indicating that TG plants have a better capacity of water retention under salt stress. The EL of WT and TG plants was similar under normal growth conditions, and then increased dramatically after NaCl treatment (Fig. 3c). However, EL in TG plants was significantly lower than that in WT controls (Fig. 3c), suggesting that TG plants have improved ability to maintain cell membrane integrity during salt exposure. Salt stress leads to chlorophyll breakdown and induces leaf yellowing. In this study, total chlorophyll content of WT and TG plants was similar before the salt stress test, but declined after salt stress exposure (Fig. 3d). However, the decline in WT controls was significantly more pronounced than that in TG plants (Fig. 3d), which may contribute to the robustness of photosynthetic system. Proline is accumulated under salinity conditions and acts as an osmolyte to protect plants from salt stress-induced toxic oxygen derivatives. Proline contents in WT and miR396 TG creeping bentgrass plants were similar under normal growth conditions, but increased dramatically after NaCl application (Fig. 3e). Interestingly, proline content in two TG lines was significantly lower than that in WT controls (Fig. 3e). Currently, the nexus between proline accumulation and abiotic stress tolerance remains controversial36. Additionally, previous study indicates that proline homeostasis is more critical than proline accumulation for the maintenance of plant growth and development under stress conditions37. Thus, it is likely that TG plants have improved capacity to maintain proline homeostasis and thereby contribute to the enhanced salt stress tolerance. It is also likely that the less salt-elicited leaf damage in TG plants triggers less accumulation of proline than WT controls.
Enhanced salt tolerance in TG plants is associated with reduced Na+ uptake
The adaptive mechanisms of plants in response to salt stress include exclusion of Na+ and/or compartmentalization of Na+ into the vacuole. To elucidate the mechanism of miR396-mediated salt tolerance, we compared the Na+ content between WT and TG plants under normal and salt stress conditions. The result shows that the two TG lines accumulate significantly less Na+ than WT controls under salt stress conditions (Fig. 4a), suggesting that the enhanced salt tolerance in miR396 TGs may be attributed to the exclusion of Na+.
a Na+ relative content and b K+ relative content were measured before and after salt stress treatment in the leaf tissue of WT and two TG lines. c K+:Na+ ratio before and after salt test in WT and two TG events. d Quantitative RT-PCR analysis of the expression levels of AsSOS1 in WT and two TG lines under normal growth conditions. AsUBQ5 was used as the reference gene. Data are shown as means (n = 3) with standard deviation. Asterisk(s) indicates significant differences between WT and each TG line at *P < 0.05; **P < 0.01; ***P < 0.001 by Student’s t-test
High concentration of Na+ during salt stress leads to limited K+ uptake and it might substitute for K+ in some K+-dependent protein interactions within plant cells. Thus, the ability of K+ retention and a high K+:Na+ ratio is important for salt tolerance. In this study, the K+ relative content is similar in WT and TG plants under both normal and stressed conditions (Fig. 4b), while the K+:Na+ ratio is significantly elevated in TG plants under salinity conditions (Fig. 4c), which might contribute to the enhanced salt tolerance.
MiR396 is involved in the regulation of Na+ transporter SOS1
The relatively low Na+ content in TG plants under salt stress conditions might result from the enhanced ability to extrude Na+ from the cytosol across the plasma membrane. SOS1 is the best-known Na+ transporter, which functions in Na+ exclusion. To reveal the molecular mechanism of miR396-mediated salt tolerance, we cloned a partial sequence of SOS1 in creeping bentgrass and analyzed the expression levels of AsSOS1 in WT and two TG lines. Quantitative RT-PCR analysis shows that AsSOS1 exhibits elevated expression in TG plants compared with WT controls (Fig. 4d), suggesting that miR396 mediates salt tolerance through Na+ excluding mechanism.
Identification of MiR396c putative targets and their responses to salt stress
To elucidate the molecular mechanism of miR396-mediated plant development and salt tolerance, we sought to identify the putative targets of miR396 in creeping bentgrass. It has been reported that the targets of miR396 encode the TFs of the GRF family. In Arabidopsis, GRF gene family contains nine members (AtGRF1–9), seven of which have miR396-binding site except AtGRF5 and AtGRF638. In rice, 12 members (OsGRF1–12) were identified, 10 of which contain miR396c target site except OsGRF11 and OsGRF1238. Expression analysis of OsGRFs in WT rice and TG rice overexpressing Osa-miR396d indicates that 9 out of 12 OsGRFs are repressed in TG plants, which are OsGRF1-8 and OsGRF1039. In this study, four GRFs (AsGRF3–6) were successfully cloned in creeping bentgrass based on the sequence of GRF homologs in rice and Brachypodium. MiR396c target sites in AsGRF3–6 were identified and compared with the target sites of rice OsGRF3–6 as shown in Fig. 5a. The expression of these four genes was repressed in TG2 and TG7 lines compared with that in WT controls (Fig. 5b), suggesting that they are the putative targets of miR396c in creeping bentgrass. GRFs play an essential role in plant leaf, stem, and root growth and development40. Therefore, it is plausible to speculate that the repressed expression of the AsGRFs might contribute to the altered plant development in miR396 TG creeping bentgrass.
a A comparison of miR396c target sites in the putative targets AsGRF3, AsGRF4, AsGRF5, and AsGRF6 between rice and creeping bentgrass. Asterisks indicate the identical RNA sequences. b Quantitative RT-PCR analysis of AsGRF3, AsGRF4, AsGRF5, and AsGRF6 expression in WT and TG plants. c Expression profiles of miR396c putative targets in response to salt stress. Transcript levels of AsGRF3, AsGRF4, AsGRF5, and AsGRF6 were analyzed at 0, 1.5, 3, and 6 h after 250 mM NaCl treatment using real-time RT-PCR. AsUBQ5 was used as an endogenous control. Data are presented as means of three technical replicates, and error bars represent ± SE. Asterisk(s) indicates a significant difference of expression levels between WT and each transgenic line at *P < 0.05; **P < 0.01; ***P < 0.001 by Student’s t-test
Next, we analyzed the expression patterns of AsGRFs under salt conditions to investigate how the miR396-GRF pathway is implicated in plant salt tolerance. Quantitative RT-PCR analysis shows that expression levels of AsGRF3–6 are all up-regulated in response to 250 mM NaCl treatment (Fig. 5c).
Genome-wide analysis of gene expression in relation to miR396-mediated plant development and salt stress tolerance
MiRNAs serve as master regulators to integrate different regulatory pathways to control plant development and plant response to environmental stress. To gain insight into the miR396-mediated regulatory network, we performed RNA-seq analysis to study the differentially expressed genes (DEGs) in miR396 TG plants vs. WT controls. Shoots RNA of non-stressed WT and TG were isolated for cDNA library preparation. Illumina sequencing generated 4,444,691 contigs, which were further assembled into 82,819 unigenes with an average size of 995.5 bp. The reproducibility of RNA-seq analysis was confirmed by multidimensional scaling (MDS) plot (Fig S3a), which shows the expected consistency between two biological replicates of WT and TG samples, respectively. A volcano plot shows the distribution of Log2-fold changes (FC) of 17,338 unigenes at false discovery rate (FDR) corrected p-value < 0.05 (Fig S3b and c). Among the differentially expressed transcripts (Log2 FC > 2 or <−2), 584 are up-regulated and 1027 are down-regulated in TG plants vs. WT controls.
Gene ontology (GO)-enrichment analysis
To identify the putative biological processes of DEGs in TG vs. WT plants, we performed GO-enrichment analysis. In this study, 9 and 12 GO terms in the biological process category were significantly enriched (over-represented P-value < 0.05) in the up-regulated and down-regulated genes (log2 FC > 2 or log2 FC < −2), respectively (Fig. S4). Among them, significantly enriched GO terms of ‘oxidation–reduction process’, ‘response to oxidative stress’, and ‘hydrogen peroxide catabolic process’ suggest that miR396 might be involved in the oxidation–reduction process. Salt stress results in ROS accumulation in plant cells and creates oxidative environment and imbalanced redox state. Oxidation–reduction process plays a fundamental role in regulating redox homeostasis and might contribute to the enhanced salt tolerance in miR396 TG plants. Besides the stress tolerance-related GO terms, GO terms of ‘DNA replication’, ‘cell division’, ‘regulation of cell cycle’, and ‘mitotic nuclear division’ were all significantly enriched in the down-regulated genes (Fig. S4b), indicating that TG plants might have less cell division and fewer cell numbers, consistent with what we observe in TG leaves.
Differential expression of leaf development-related and stress response-related genes
In addition to identify the overrepresented GO terms in the whole DEGs of TG vs. WT dataset, we also analyzed the leaf development-related and stress response-related DEGs. Significantly enriched GO terms (over-represented P-valueP < 0.05) in the DEGs (log2 FC > 1 or log2 FC < −1, FDR-corrected P-value < 0.05) were identified, in which the corresponding genes related to leaf development and environmental stress response were chosen for generating a heatmap. As shown in Fig. S5, 201 DEGs had enriched GO terms of ‘DNA replication’, ‘cell differentiation’, ‘cell division’, ‘leaf development’, etc. In addition, 603 DEGs had enriched GO terms related to biotic and abiotic stress responses, including response to cold, salt, heat, drought, wounding, light, heavy metal, nutrient deficiency, bacterium, virus, and fungus (Fig. S5). The result demonstrates that miR396 affects a variety of biological processes.
Validation of expression profiles of candidate genes
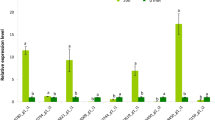
To validate the expression profiles of DEGs in RNA-seq data, we analyzed expression levels of candidate genes involved in cell reproduction and environmental stress response (Fig. 6). As1053 is predicted to encode a subunit of anaphase-promoting complex involved in cell reproduction. RT-PCR analysis shows repressed expression in two TG lines compared with that in WT controls (Fig. 6), in agreement with RNA-seq result. Also consistent with the RNA-seq data, a predicted WRKY40 transcription factor gene, As71896 and two MADS-box genes, As37603 and As40994 are up-regulated in TG plants in comparison with WT controls (Fig. 6). Furthermore, As3793, a predicted AP2 transcription factor gene, is down-regulated in both RT-PCR and RNA-seq results (Fig. 6).
Discussion
MiR396-mediated plant development during vegetative growth
The miR396-GRF regulatory pathway plays a key role in leaf development. Overexpression of At-miR396a leads to smaller and narrower leaves through suppressing GRF genes in TG tobacco20. In this study, TG creeping bentgrass overexpressing Os-miR396c resulted in similar leaf phenotype and down-regulated AsGRFs, implying that miR396-GRF pathway is functionally conserved in different plant species.
The role of GRFs in regulating leaf development has been well characterized in Arabidopsis. The quadruple mutant atgrf1/2/3/4 exhibits smaller leaf size because of the reduced cell size and cell number40. AtGRF1 and AtGRF2 control cell expansion, while AtGRF3 and AtGRF4 regulate cell proliferation40. In this study, we compared the number of leaf epidermis cells per unit leaf area between WT and TG plants. The result shows that TG plants have significantly fewer leaf epidermis cells per unit area than that of WT controls, which also implies larger cell size in TG plants. Additionally, RT-PCR analysis shows that As1053, a putative gene encoding anaphase-promoting complex subunit, which is involved in cell proliferation, is down-regulated in miR396 TG plants. Thus, it is likely that miR396c/GRF module control leaf area through regulating cell proliferation instead of cell expansion. This is further supported by the GO-enrichment analysis, in which the GO terms of ‘DNA replication’, ‘cell division’, ‘regulation of cell cycle’, and ‘mitotic nuclear division’ were significantly enriched in the down-regulated genes.
In grasses, tillering plays an essential role in shoot architecture and grain yield. In this study, tillering differences were observed between WT and miR396 TG creeping bentgrass. TG plants accumulate less shoot biomass, and have fewer tillers with reduced tiller length due to shorter internodes, suggesting that miR396 is implicated in the regulation of tillering. The development of grass tillers is determined by shoot apical meristems (SAMs), since all aerial organs are produced from SAMs. Previous studies show that KNOTTED1-LIKE HOMEOBOX (KNOX) transcription factor genes are key controllers on SAM formation and maintenance41,42. Recently, GRFs have been reported to regulate the expression of KNOX family gene OsKN2 in rice through interacting with the promoter of OsKN243. The similar interactions between GRF and KNOX have also been reported in Arabidopsis and barley43,44. Therefore, we speculate that the altered tillering in TG creeping bentgrass may be mediated through the miR396-GRF-KNOX regulatory module.
MiR396 is also involved in the modulation of root development. TG Medicago truncatula overexpressing miR396b exhibits reduced root dry weight, while MIM396, the inactive form of miR396 TG line displays significantly increased root dry weight compared with WT controls45. Further analysis indicates that miR396 modulates root growth through restricting the activity of root cell division43. In this study, the phenotype of reduced root dry weight is also observed in miR396 TG creeping bentgrass, indicating the conserved function of miR396-mediated root development in both monocots and dicots.
MiR396-mediated regulatory network in plant response to salt stress
Plant responses to high salinity conditions have been extensively studied at the molecular level. The products of high salinity-responsive genes include downstream functional proteins and upstream regulatory proteins. In this study, AsSOS1, encoding the downstream functional Na+/H+ antiporter, was up-regulated in miR396 TG plants. Under salinity stress, SOS1 functions in maintaining ion homeostasis. Previous studies show that overexpression of SOS1 leads to enhanced salt tolerance in TG Arabidopsis, tobacco, and Chrysanthemum crassum11,46,47. It is plausible that the enhanced salt tolerance in TG creeping bentgrass may result from miR396-mediated positive regulation of AsSOS1.
In addition, we also investigated the potential role other important Na+ transporters, NHX and HKT may play in miR396-mediated plant stress tolerance. NHX transporters are one of the best-characterized Na+ transporters, which alleviate excess Na+ through mediating intracellular Na+/H+ and K+/H+ antiport to compartmentalize Na+ into the vacuoles5. To examine the possible involvement of NHX in miR396-mediated plant response to salt stress in creeping bentgrass, a rice NHX gene OsNHX1 was used to blast against the creeping bentgrass RNA-seq data to identify and clone the AsNHX gene. Real-time PCR analysis of AsNHX expression revealed no significant difference between miR396 TG and WT control plants (Figure S6a), suggesting that NHX may not be involved in miR396-mediated plant salt stress tolerance in miR396 TG creeping bentgrass. Our results indicate that the enhanced salt tolerance in miR396 TG plants is associated with reduced Na+ uptake, which suggests the adaptive mechanism of Na+ exclusion instead of Na+ compartmentalization into the vacuole in TG plants.
HKT transporters exhibit species-specific function due to their different selectivity for Na+ and K+. In wheat, TaHKT2;1 showed Na+/K+ co-transport activity at high Na+ as compared to K+ in extracellular compartment48,49. In T. salsuginea, a plant closely related to Arabidopsis, TsHKT1;2 showed higher selectivity for K+ over Na+50. In Arabidopsis, AtHKT1;1 showed a strong Na+ selective transport activity. AtHKT1 may participate in the Na+ uptake and Na+ circulation in plant body51. One of the rice HKTs exhibited the similar function52. To study the possible role of HKT in miR396-mediated plant salt tolerance in creeping bentgrass, a rice gene OsHKT1 was used to blast against RNA-seq data in creeping bentgrass and identify AsHKT1 for further study. Quantitative RT-PCR analysis of AsHKT1 revealed a significantly reduced gene expression in miR396 TG creeping bentgrass compared with WT controls (Figure S6b), suggesting its possible involvement in the miR396-mediated plant salt stress response pathway. It is plausible that AsHKT1 may have higher selectivity for Na+ than K+. Overexpression of miR396 negatively impacts AsHKT1 expression, leading to a reduced accumulation of Na+ in TG plant cells under salt stress conditions. The exact role AsHKT1 plays in miR396-mediated salt stress response in creeping bentgrass remains to be further explored in the future.
Besides ionic stress, high salinity leads to the production of ROS, which damages plant cells and tissues through interacting with key macromolecules and metabolites. Among functional proteins, antioxidant enzymes protect plants from oxidative stress caused by ROS accumulation. In this study, significantly enriched GO terms of ‘oxidation–reduction process’, ‘H2O2 catabolic process’, ‘response to oxidative stress’, ‘oxidoreductase activity’, and ‘peroxidase activity’ imply that miR396 is crucial in plant response to oxidative stress.
Upstream regulatory proteins involved in high salinity responses include TFs, protein kinases, and phosphatases. The GO-enrichment analysis shows that ‘regulation of transcription’ and ‘regulation of kinase activity’ are significantly enriched in the down-regulated genes (Fig. S4b). Further RT-PCR analysis shows that genes encoding TFs from WRKY, MADS, and AP2 families are induced or down-regulated in miR396 TG plants (Fig. 6). TFs from WRKY family positively regulate salt and other environmental stress tolerance in a variety of plant species53,54. MADS and AP2 family TFs are also largely involved in plant abiotic stress resistance55,56. In addition to salt stress response, significantly enriched DEGs also participate in the biological processes of response to heat, cold, drought, heavy metal, wounding, high light intensity, nutrient deficiency stresses, and defense response to bacterium, virus, and fungus. Therefore, the result suggests that miR396, in a broad sense, may be actively involved in multiple environmental stress responses through modulating both functional and regulatory proteins.
MiR396 exerts its function at post-transcriptional and post-translational levels to regulate its targets, which encode TFs of GRF family and some species-specific TFs (e.g. bHLH79 in Medicago, SVP in Arabidopsis)45,57. These TFs will further regulate their direct targets, including TFs or other regulatory and functional proteins (e.g. AtGRF7 targets TF gene DREB2A in Arabidopsis, OsGRF3 and OsGRF10 target TF gene OsKN2 in rice, OsGRF6 and OsGRF10 target protein kinase gene OsCR4 and demethylase gene OsJMJ706)39,58,59. Therefore, miR396 serves as an important master regulator to integrate various regulatory elements to help plants cope with salt and other environmental stresses.
Participation of GRFs in plant abiotic stress responses
In Arabidopsis, one of the miR396 target genes AtGRF7 functions as a repressor of stress-responsive genes in plant abiotic stress response59. Under non-stressed conditions, AtGRF7 overexpression lines and atgrf7 mutants showed similar phenotype to WT controls. Interestingly, AtGRF7 knock out mutants exhibited salt and drought tolerance in comparison with WT controls and AtGRF7 overexpression lines. Further study showed that AtGRF7 suppressed DREB2A (DEHYDRATION-RESPONSIVE ELEMENT BINGING PROTEIN2A) expression through biding to its promoter59. In addition, analysis of the T-DNA insertion mutant, atgrf7-1 revealed a large number of up-regulated genes that are involved in stress and abscisic acid responses, including DREB2A59. In our RNA-seq results, expression of AsDREB2A is also elevated in miR396 TG creeping bentgrass compared to wild type controls, which suggests that miR396-GRF7-DREB2A signaling pathway might play an important role contributing to the enhanced salt tolerance in TGs.
Taken together, the data obtained in our study suggest a potential signaling pathway of miR396-mediated salt tolerance in creeping bentgrass (Fig. 7). In response to high salinity, miR396 is induced to post-transcriptionally control the regulatory proteins of protein kinases and TFs, such as GRFs, DREB2A, and WRKY40. Next, these regulatory proteins activate downstream salinity responsive genes, such as Na+/H+ antiporter, AsSOS1; detoxification enzyme genes to protect plant cells from ionic stress and oxidative stress caused by high salinity stress; and DREB2A target genes to protect macromolecules from dehydration. This study provides insight into miR396-mediated salt tolerance and allows deciphering the role of miRNAs in the complex regulatory network in plant response to environmental stress.
References
Hasegawa, P. M., Bressan, R. A., Zhu, J. & Bohnert, H. J. Plant cellular & molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499 (2000).
Munns, R. & Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008).
Uozumi, N. & Schroeder, J.I. Ion channels and plant stress: past, present, and future. In Ion Channels and Plant Stress Responses (eds. Demidchik, V., Maathuis, F.) 1–22 (Springer, Berlin, 2010).
Hauser, F. & Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+ /Na+ ratio in leaves during salinity stress. Plant Cell Environ. 33, 552–565 (2010).
Blumwald, E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12, 431–434 (2000).
Zhang, H. X., Hodson, J. N., Williams, J. P. & Blumwald, E. Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl Acad. Sci. USA 98, 12832–12836 (2001).
Agarwal, P. K., Shukla, P. S., Gupta, K. & Jha, B. Bioengineering for salinity tolerance in plants: state of the art. Mol. Biotechnol. 54, 102–123 (2013).
Wu, S. J., Ding, L. & Zhu, J. K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8, 617–627 (1996).
Quan, R. et al. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19, 1415–1431 (2007).
Quintero, F. J. et al. Activation of the plasma membrane Na+/H+ antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl Acad. Sci. USA 108, 2611–2616 (2011).
Yang, Q. et al. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2, 22–31 (2009).
Ben Ahmed, C., Ben Rouina, B., Sensoy, S., Boukhriss, M. & Ben Abdullah, F. Exogenous proline effects on photosynthetic performance and antioxidant defense system of young olive tree. J. Agric. Food Chem. 58, 4216–4222 (2010).
Garg, A. K. et al. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl Acad. Sci. USA 99, 15898–15903 (2002).
Sharma, P., Jha, A. B., Dubey, R. S. & Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot., Article 217037. https://doi.org/10.1155/2012/217037 (2012).
Eltayeb, A. E. et al. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225, 1255–1264 (2007).
Guan, Z., Chai, T., Zhang, Y., Xu, J. & Wei, W. Enhancement of Cd tolerance in transgenic tobacco plants overexpressing a Cd-induced catalase cDNA. Chemosphere 76, 623–630 (2009).
Ma, C., Burd, S. & Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 84, 169–187 (2015).
Yuan, S. et al. Constitutive expression of rice microRNA528 alters plant development and enhances tolerance to salinity stress and nitrogen starvation in creeping bentgrass. Plant Physiol. 169, 576–593 (2015).
Kim, J. H., Choi, D. & Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104 (2003).
Yang, F., Liang, G., Liu, D. & Yu, D. Arabidopsis miR396 mediates the development of leaves and flowers in transgenic tobacco. J. Plant Biol. 52, 475–481 (2009).
Rodriguez, R. E. et al. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112 (2010).
Baucher, M. et al. A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol. 15, 892–898 (2013).
Debernardi, J. M. et al. Post‐transcriptional control of GRF transcription factors by microRNA miR396 and GIF co‐activator affects leaf size and longevity. Plant J. 79, 413–426 (2014).
Casadevall, R., Rodriguez, R. E., Debernardi, J. M., Palatnik, J. F. & Casati, P. Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell 25, 3570–3583 (2013).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Method gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010).
Chen, L., Luan, Y. & Zhai, J. Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 34, 2013–2025. https://doi.org/10.1007/s00299-015-1847-0. (2015).
Gao, P. et al. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 231, 991–1001 (2010).
Xiao, H. et al. Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol. Biol. 52, 957–966 (2003).
Luo, H., Hu, Q., Nelson, K., Longo, C. & Kausch, A.P. Controlling Transgene Escape in Genetically Modified Grasses. In: Hopkins, A., Wang, Z.Y., Mian, R., Sledge, M., & Barker, R.E. (eds) Molecular Breeding of Forage and Turf. Developments in Plant Breeding, Vol. 11. 245–254 (Springer, Dordrecht, 2004).
Luo, H. et al. Agrobacterium tumefaciens-mediated creeping bentgrass (Agrostis stolonifera L.) transformation using phosphinothricin selection results in a high frequency of single-copy transgene integration. Plant Cell Rep. 22, 645–652 (2004).
Li, Z., Baldwin, C. M., Hu, Q., Liu, H. & Luo, H. Heterologous expression of Arabidopsis H‐pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (Agrostis stolonifera L.). Plant Cell Environ. 33, 272–289 (2010).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Liao, Y., Smyth, G. K. & Shi, W. Feature Counts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Zhou, J. et al. Expression profile of miRNAs in Populus cathayana L. and Salix matsudana Koidz under salt stress. Mol. Biol. Rep. 39, 8645–8654 (2012).
Qin, Z., Chen, J., Jin, L., Duns, G. J. & Ouyang, P. Differential expression of miRNAs under salt stress in Spartina alterniflora leaf tissues. J. Nanosci. Nanotechnol. 15, 1554–1561 (2015).
Hare, P. D. & Cress, W. A. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 21, 79–102 (1997).
Kishor, K. P. B. & Sreenivasulu, N. Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ. 37, 300–311 (2014).
Debernardi, J. M., Rodriguez, R. E., Mecchia, M. A. & Palatnik, J. F. Functional specialization of the plant miR396 regulatory network through distinct microRNA–target interactions. PLoS Genet. 8, e1002419 (2012).
Liu, H. et al. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 165, 160–174 (2014).
Omidbakhshfard, M. A., Proost, S., Fujikura, U. & Mueller-Roeber, B. Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010 (2015).
Long, J. A., Moan, E. I., Medford, J. I. & Barton, M. K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69 (1996).
Vollbrecht, E., Reiser, L. & Hake, S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127, 3161–3172 (2000).
Kuijt, S. J. et al. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164, 1952–1966 (2014).
Osnato, M. et al. Cross talk between the KNOX and ethylene pathways is mediated by intron-binding transcription factors in barley. Plant Physiol. 154, 1616–1632 (2010).
Bazin, J. et al. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 74, 920–934 (2013).
An, J. et al. The over-expression of Chrysanthemum crassum CcSOS1 improves the salinity tolerance of chrysanthemum. Mol. Biol. Rep. 41, 4155–4162 (2014).
Yue, Y., Zhang, M., Zhang, J., Duan, L. & Li, Z. SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. Plant Physiol. 169, 255–261 (2012).
Rubio, F., Gassmann, W. & Schroeder, J. I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270, 1660–1663 (1995).
Gassmann, W., Rubio, F. & Schroeder, J. I. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 10, 869–882 (1996).
Ali, Z. et al. TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol. 158, 1463–1474 (2012).
Yamaguchi, T., Hamamoto, S. & Uozumi, N. Sodium transport system in plant cells. Front. Plant Sci. 4, 410 (2013).
Ren, Z. H. et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146 (2005).
Wang, X. et al. Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 6, 615 (2015).
Zhao, T. et al. Expression and functional analysis of WRKY transcription factors in Chinese wild hazel, Corylus heterophylla Fisch. PLoS ONE 10, e0135315 (2015).
Arora, R. et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 8, 242 (2007).
Akhtar, M. et al. DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J. Genet. 91, 385–395 (2012).
Yang, C. Y. et al. MicroRNA396-targeted SHORT VEGETATIVE PHASE is required to repress flowering and is related to the development of abnormal flower symptoms by the Phyllody Symptoms1 Effector. Plant Physiol. 168, 1702–1716 (2015).
Kuijt, S. J. et al. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164, 1952–1966 (2014).
Kim, J. S. et al. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24, 3393–3405 (2012).
Acknowledgements
This project was supported by Biotechnology Risk Assessment Grant Program competitive grant no. 2010-33522-21656 from the USDA National Institute of Food and Agriculture, as well as the USDA grant CSREES SC-1700450. Technical Contribution No. 6289 of the Clemson University Experiment Station.
Author information
Authors and Affiliations
Contributions
S.Y., J.Z, S.L., and H.L. conceived the project, designed the experiments, and analyzed the data; S.Y., J.Z., Z.L., Q.H., N.Y., M.Z., and Q.H. carried out the experiments; X.X., R.N., and C.S. contributed to RNA-seq data analysis; S.Y., J.Z., and H.L. wrote the article.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yuan, S., Zhao, J., Li, Z. et al. MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Hortic Res 6, 48 (2019). https://doi.org/10.1038/s41438-019-0130-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-019-0130-x
This article is cited by
-
AsHSP26.2, a creeping bentgrass chloroplast small heat shock protein positively regulates plant development
Plant Cell Reports (2024)
-
Genome-wide analysis of long noncoding RNAs in response to salt stress in Nicotiana tabacum
BMC Plant Biology (2023)
-
A conserved SNP variation in the pre-miR396c flanking region in Oryza sativa indica landraces correlates with mature miRNA abundance
Scientific Reports (2023)
-
Endoplasmic reticulum stress-responsive microRNAs are involved in the regulation of abiotic stresses in wheat
Plant Cell Reports (2023)
-
Enhancing alfalfa resistance to Spodoptera herbivory by sequestering microRNA396 expression
Plant Cell Reports (2023)