Abstract
Correctly determining species’ identity is critical for estimating biodiversity and effectively managing marine populations, but is difficult for species that have few morphological traits or are highly plastic. Sponges are considered a taxonomically difficult group because they lack multiple consistent diagnostic features, which coupled with their common phenotypic plasticity, makes the presence of species complexes likely, but difficult to detect. Here, we investigated the evolutionary relationship of Tethya spp. in central New Zealand using both molecular and morphological techniques to highlight the potential for cryptic speciation in sponges. Phylogenetic reconstructions based on two mitochondrial markers (rnl, COI-ext) and one nuclear marker (18S) revealed three genetic clades, with one clade representing Tethya bergquistae and two clades belonging to what was a priori thought to be a single species, Tethya burtoni. Eleven microsatellite markers were also used to further resolve the T. burtoni group, revealing a division consistent with the 18S and rnl data. Morphological analysis based on spicule characteristics allowed T. bergquistae to be distinguished from T. burtoni, but revealed no apparent differences between the T. burtoni clades. Here, we highlight hidden genetic diversity within T. burtoni, likely representing a group consisting of incipient species that have undergone speciation but have yet to express clear morphological differences. Our study supports the notion that cryptic speciation in sponges may go undetected and diversity underestimated when using only morphology-based taxonomy, which has broad scale implications for conservation and management of marine systems.
Similar content being viewed by others
Introduction
Correctly identifying species units is critical for estimating biodiversity, defining species boundaries and measuring connectivity patterns (Hey et al. 2003; Frankham et al. 2012). Methods for species identification have been so hotly debated in the literature that the phrase ‘species problem’ has emerged to describe the conundrum that biologists face when defining a species (Mayden 1997; De Queiroz 2007). Mayr (1942) was the first to address this problem by conceiving the biological species concept, where he defined a species as a group of interbreeding individuals which are reproductively isolated from other groups. Since then, other concepts have emerged based on: an organism’s functional role/ecological niche (ecological species concept, Van Valen 1976); historical ancestry lineages (evolutionary species concept, Simpson 1951); and the sharing of common ancestors/alleles (phylogenetic species concept, Hennig 1966; Rosen 1979; Baum and Shaw 1995; reviewed in De Queiroz 2007). While many concepts exist, the most traditional, widespread and consistently employed method for taxonomy is based on comparative morphology (Padial et al. 2010).
The recent advancement and accessibility of molecular tools has begun to reveal conflicting evolutionary histories for many species previously defined by morphological characteristics (Wendel and Doyle 1998; Knowlton 2000; Lecompte et al. 2005; Lihová et al. 2007; Wallis et al. 2017). Distinguishing between intraspecific plasticity and interspecific cryptic diversity can be problematic when only using morphology-based taxonomic methods (Knowlton 1993; Vrijenhoek 2009; Capa et al. 2013, Czarna et al. 2016). Additionally, congeneric species that live in sympatry often challenge the notion of species boundaries because introgression (i.e. where alleles from one species enter the gene pool of another species via hybridization and backcrossing) is common (Harrison and Larson 2014). Failure to correctly identify species boundaries has consequences for implementing effective conservation and management strategies and for our overall understanding of important evolutionary processes like speciation (Gómez et al. 2002), bioinvasions (Knapp et al. 2015) and hybridization (Kauserud et al. 2006).
Organisms that have only a few defining traits available for comparison are particularly prone to identification problems (Knowlton 1993; Bickford et al. 2007; DeBiasse and Hellberg 2015). In such groups, cryptic species often emerge during molecular studies, as sequences provide more characteristics (bases) with which to differentiate organisms, which allows a finer resolution of relatedness between individuals to be determined (Hilis 1987; Hebet et al. 2004; Bickford et al. 2007). Sponges have relatively few morphological traits, yet a rich taxonomic literature exists delineating species boundaries within sponges based on phenotypic characteristics (Hooper and Van Soest 2002). Skeletal composition, arrangement and spicule morphology are key features used to classify and identify sponges (Hooper and Van Soest 2002), despite the fact that structural components of sponges can be highly dynamic (Bond 1992; Nakayama et al. 2015) and spicules can exhibit variability under different environmental conditions (Bavestrello et al. 1993; Bell et al. 2002; McDonald et al. 2002). Furthermore, recent studies on sponge taxonomy have highlighted problems with using morphology as a sole defining trait and have used phenotypic traits combined with molecular analyses to resolve evolutionary relationships (Alvarez et al. 2000; Erwin and Thacker 2007; Morrow and Cárdenas 2015; Plotkin et al. 2017). As a result, phylogenetic analyses for many sponges have begun to reveal cryptic speciation throughout the phylum, suggesting that diversity within the phylum is even higher than previously thought (see Table 1 in Xavier et al. (2010) for reported cases of cryptic speciation in sponges, plus Andreakis et al. 2012; De Paula et al. 2012; DeBiasse and Hellberg 2015; Knapp et al. 2015; Uriz et al. 2017).
In this study, we used ‘golf ball sponges’ belonging to the genus Tethya in New Zealand as a model to investigate cryptic speciation and to better understand how evolutionary processes occur for species living in sympatry. Tethya is a ubiquitous genus, with 92 described species worldwide (Van Soest 2008), and has a particularly high diversity (31 species) recorded in coastal waters of Australia and New Zealand (Bergquist and Kelly-Borges 1991; Sarà 1998; Sarà and Sarà 2004; Heim et al. 2007). While members of the genus are easily distinguishable by their globular form, identifying Tethya to the species level can be difficult because they exhibit a high degree of morphological plasticity, and have only a few defining traits available for species differentiation. The challenge in species identification is reflected in the dynamic taxonomic literature, where new species are constantly being described (Corriero et al. 2015), and previously described species are regularly reconsidered and reclassified (Sarà 1987; Bergquist and Kelly-Borges 1991; Hooper and Wiedenmayer 1994; Sarà and Sarà 2004; Heim et al. 2007). In New Zealand, taxonomy has been based solely on morphology with no genetic phylogenies constructed to date (Bergquist and Kelly-Borges 1991; Sarà and Sarà 2004). Here, we used two common species of Tethya in New Zealand (Tethya bergquistae and Tethya burtoni) as models to investigate evolutionary relationships and cryptic speciation using both genetic (COI-ext, rnl, 18S and 11 microsatellite markers) and morphological traits (spicules) in order to understand the advantages and limitations of both methods in determining species boundaries.
Materials and methods
Study species
Tethya bergquistae and T. burtoni are two of the most common species of Tethya found in the waters of central New Zealand, and both occur in the waters around Wellington, New Zealand (Fig. 1). Tethya bergquistae is described as rose-pink in colour, firm to the touch and with an irregular surface with grouped oscula (Bergquist and Kelly-Borges 1991; Hooper and Wiedenmayer 1994; Battershill et al. 2010). Tethya burtoni is bright orange to yellow and is described as having a warty surface with an inflated, soft texture (Sarà and Sarà 2004; Battershill et al. 2010). Tethya bergquistae is generally found in well-lit areas with moderate flow, whereas T. burtoni tends to prefer more shaded, sedimented areas (Battershill et al. 2010). Spicules for both include megascleres, which are strongyloxeas; and microscleres, which are megasters and micrasters of similar sizes (Bergquist and Kelly-Borges 1991; Sarà and Sarà 2004). A key distinguishing feature between both species relies on their spicular composition with respect to the location/density of megasters, as T. burtoni contains fewer megasters, which are mainly found in the cortex (Bergquist and Kelly-Borges 1991; Sarà and Sarà 2004).
Examples of sponges collected belonging to clades identified in Fig. 3, to allow comparison of macromorphological features. For each clade, cross sections that encompass the ectosome and choanosome are included for a subsample (n = 3) of genetically identical individuals. Clade 1 (top) are examples of Tethya bergquistae, while Clades 2 and 3 (middle and bottom, respectively) are examples of T. burtoni. Scale bars in each photo = 10 mm
Tracking the description of both species through the literature reveals a complex taxonomic classification history within New Zealand, highlighting the complexity in species identification common to Porifera. For instance, T. bergquistae was incorrectly identified as Tethya ingalli by Pritchard et al. (1984) and formerly described as Tethya australis by Bergquist and Kelly-Borges (1991), and T. burtoni has been also incorrectly referred to as Tethya aurantium by Pritchard et al. (1984). Overall in New Zealand, there are ten species of Tethya recorded, but two of these are also thought to exist in Australian waters (Ledenfield 1888; Bergquist 1961; Bergquist and Kelly-Borges 1991; Sarà and Sarà 2004). Many of these ten species are thought to be endemic to certain regions (e.g. Tethya compacta to the Chatham Islands and Tethya fastigata to the Poor Knight Islands), but this has been based on observations from only one to a few sponges (e.g. T. compacta), or on descriptions dating from the 19th Century (e.g. Tethya multistella). In the literature, pictures identifying New Zealand Tethya are often inconsistent and classification by comparative morphology is therefore challenging. Descriptions of species within Tethya are based on cross sections and spicule compliments, but differences can be often subtle and difficult to detect, especially for non-specialists. Around New Zealand, sympatric Tethya spp. display a wide range of colours and textures, making them difficult to confidently match to descriptions in the current taxonomic literature. Tethya also appear to undergo seasonal morphological changes (M. Shaffer, personal observations from monthly monitoring for 2+ years). For example, the same sponge’s appearance may go from porous and soft to very firm over a period of a month. In addition, during the summer a sponge may be covered more extensively by crustose coralline algae (CCA), rendering it more reminiscent of Tethya amplexa (Bergquist and Kelly-Borges 1991), but in the winter be completely free of CCA. Because morphology can be very variable and temporally dependent, assessing species based solely on morphology has the potential to lead to erroneous conclusions. To date, molecular differences have allowed at least one other instance of sibling species within the genus Tethya to be uncovered. Sarà et al. (1993) reported cryptic speciation in two populations of Tethya robusta, which were morphologically similar but genetically different. The taxonomic history, phenotypic plasticity and previous report of a species complex within the Tethya genus makes other occurrences of cryptic speciation within this group likely.
Collection
Sponges which were a priori thought to be T. bergquistae and T. burtoni were collected using SCUBA from two different locations in waters around Wellington, New Zealand (Breaker Bay: 41°19′53.3″S 174°49′52.6″E; Kapiti Island: 40°53′23.6″S 174°52′40.3″E). Additional specimens of T. burtoni were collected from two further locations (Somes Island: 41°15′36.9″S 174°43′54.4″E; Red Rocks: 41°21′04.5″S 174°43′54.4″E). Sponges were collected 3–5 m apart to avoid collecting clones, at depths from 5 to 10 m. Three sponges of unknown identity were collected from Breaker Bay (n = 2) and Kapiti Island (n = 1), which were texturally similar to T. bergquistae but the same colour as T. burtoni. A picture of each specimen was taken to aid in later identification. Tissue was extracted for both genetic and spicule analysis, and the rest of the specimen was preserved in ethanol. In total, 71 sponges of various morphologies were collected for phylogenetic and morphological analyses. For those sponges assumed to be T. bergquistae, samples were collected from Breaker Bay (n = 10) and Kapiti Island (n = 5). For those sponges assumed to be T. burtoni, samples were collected from Breaker Bay (n = 17), Kapiti Island (n = 20), Red Rocks (n = 5) and Somes Island (n = 12). For information on specimens collected for this study, see Table S1 in the Supplementary Information.
DNA extraction, PCR amplification, sequencing and fragment analysis
Tissue used for genetic analysis was taken from the inside (choanosome) of the sponge and rinsed thoroughly to reduce the risk of contamination from epibionts or other associated organisms. DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen), following the protocol of the manufacturer. We tried five primer sets, but two regions failed to consistently give readable sequences across all samples (ITS, primer set: ITS2.2/ITSRA2, as in Adlard and Lester 1995; and COI, primer set: dgLCO1490/dgHCO2198; Folmer et al. 1994), so were discarded for further analysis. Three markers were amplified in total: 18S, rnl and COI-ext. 18S is a nuclear gene that codes ribosomal DNA (rDNA), and rnl is a mitochondrial partition that codes the large ribosomal subunit RNA (Boore 1999). COI-ext is a region downstream of COI, which is a mitochondrial coding region for the cytochrome c oxidase subunit used in cellular metabolism (Folmer et al. 1994). This extended region is thought to be more informative for closely related species because its substitution pattern (mainly consisting of transversions) reveals a more progressive stage of character evolution (Erpenbeck et al. 2006a). As such, this region exhibits variation during early stages of species divergence compared to the traditionally conserved COI region (by Folmer et al. 1994) that is commonly used (Erpenbeck et al. 2006a). Primer information and product sizes are shown in Table 1.
Reaction and cycling conditions were similar for all three markers. Reactions were carried out in 30 µl volumes consisting of: 15 µl MyTaq Red Mix (Bioline), 0.6 µl of each primer (10 μM), ~25 ng template DNA and a volume of distilled water to reach 30 µl. Amplification profiles were as follows: an initial denaturation at 94 °C for 5 min; 35 cycles of 94 °C for 30 s, Ta for 55 s (Ta = 50 °C for 18S; Ta = 58 °C for COI-ext and rnl), 72 °C for 45 s; followed by a final extension at 72 °C for 7 min. Products were visually checked on a 1.5% agarose gel stained with RedSafe Nucleic Acid Staining Solution (20,000×) and sequenced using automated DNA sequencing services provided by Macrogen, Inc. (Seoul, South Korea). Sequences were deposited into GenBank, with accession numbers for haplotypes of all individuals provided in Table S1 in the Supplementary Information.
Eleven microsatellite markers were developed for T. burtoni (see Supplementary Information for microsatellite development and Table S2 for microsatellite information). These markers failed to amplify for T. bergquistae. Fragment analysis was performed on sponges belonging to the T. burtoni clades. Reactions were carried out in 30 µl reactions, with 15 µl MyTaq Red Mix (Bioline), 0.6 µl of each primer (10 μM), ~25 ng template DNA and a volume of distilled water to reach 30 µl. Amplification profiles were as follows: an initial denaturation at 94 °C for 10 min; 35 cycles of 94 °C for 45 s, 60 °C for 55 s, 72 °C for 60 s; followed by a final extension at 72 °C for 10 min. Fluorescently labelled products were visually checked, pooled, purified and then genotyped using the 3730xl DNA Analyzer (Applied Biosystems) from GeneScan Services for fragment analysis by Macrogen Inc. (Seoul, South Korea). Alleles were visualized in GeneMarker v2.2 (Hulce et al. 2011) and scored manually relative to a size standard (LIZ500). Some sponges that were thought to be T. burtoni had a lower amplification success for some loci, even after redoing the DNA extraction and fragment analysis two to three times. Because of this, missing information was considered informative in analyses.
Genetic analysis
All sequences were checked for contaminants and verified as being poriferan using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences were edited in Geneious 11.0.2 (Kearse et al. 2012) and aligned using CLUSTALW2.0 (Larkin et al. 2007), as implemented in Geneious 11.0.2 (Kearse et al. 2012). Phylogenetic analyses were applied to 18S, rnl and COI-ext separately, as well as to the mitochondrial partitions concatenated (COI-ext + rnl), creating four data sets. MrModeltest v2.3 (Nylander 2004) was used to select the best-fitting model of nucleotide substitution for each data set under the Akaike information criterion, and it was determined that the SYM + G model was the best fitting model for all data sets. The closely related Tethya actinia was chosen as an outgroup because both the 18S region and its complete mitochondrion genome have been sequenced and made available on GenBank, making it an ideal candidate to root all three markers (accession numbers: AY320033 for rnl and COI-ext; AY878079 for 18S).
Phylogenetic trees were constructed using both Maximum Likelihood (ML) and Bayesian Inference (BI) methods for each data set. ML constructions were performed in PAUP* 4.0 (Swofford 2002), and BI constructions were performed in MrBayes 3.2.6 (Ronquist et al. 2012), using the model criterion generated from MrModeltest v2.3 (Nylander 2004). For ML analyses, a bootstrap analysis using a tree-bisection-reconnection (TBR) heuristic search with 1000 replicates was performed to determine topological confidence of the tree. For BI analyses, posterior probability of trees was estimated using Monte Carlo Markov Chain (MCMC) analyses, where one million generations were run sampling four chains every 100 generations, with a burn-in of 0.25. For each marker, p distances were calculated within and between clades as a measure of divergence using MEGA 7 (Kumar et al. 2015). In addition, p distances were calculated between and within T. bergquistae (Clade 1) and T burtoni (Clades 2 + 3, combined).
For microsatellite data from sponges belonging to the T. burtoni clades, a principal coordinate analysis (PCA) based on genetic distances from allele frequencies was performed in the adegenet package (Jombart 2008) in R (https://www.r-project.org/). Assignment of individuals into genetic clusters was also assessed using STRUCTURE, which is a Bayesian clustering analysis for multilocus genotype data which probabilistically assigns individuals to clusters (Pritchard et al. 2000). Analyses were carried out in STRUCTURE v 2.3.2 (Pritchard et al. 2000), using 100,000 Markov Chain Monto Carlo (MCMC) iterations, a burn in of 10,000 iterations, 10 replicates per run and with the K value set from 1 to 8. The optimal K was determined in Structure Harvester (Earl and vonHoldt 2012) to be K = 4, and the 10 replications for K = 4 were merged in CLUMPP (Jakobsson and Rosenberg 2007). Plots were visualized in MS Excel (2016). Clusters within T. burtoni were consistent with phylogenetic clades produced by 18S and rnl, and the degree of differentiation between these two groups was assessed using Fst. Because individuals belonging to Clade 3 contained loci that failed to amplify (missing data), these loci were removed for calculations of Fst. A pairwaise Fst value was calculated from the remaining 5 loci with significance determined by performing 20,000 permutations in Arlequin 3.1 (Excoffier et al. 2005). In addition, because measures of differentiation based on distance are sensitive to deviations from Hardy Weinberg Equilibrium (HWE; Waples 2015), Fst was calculated again removing one (out of five) loci that was not in HWE (Supplementary Information).
Morphological analysis
To determine if morphological characteristics were consistent with the observed genetic clades, a subsample of six sponges from each clade was selected for spicule analysis. We chose genetically identical individuals that fell consistently into the same clade for all three markers. Samples were collected from the same location and at the same time in order to reduce potential environmental and seasonal variation in spicule morphology (Bell et al. 2002). Each sponge was cut in half to photograph its cross section. Further, a small piece of tissue was embedded in paraffin wax and sectioned to 20 μm using a rotary microtome (Leica Biosystems RM2235) to examine the location, arrangement and density of spicules across the ectosome and choanosome. A ~5 mm3 tissue sample was then removed from each specimen and dissolved in bleach at 50 °C for 48 h and the remaining spicules were washed with distilled water and rinsed 3× with absolute ethanol, following the recommendations of Hooper (2003). One millilitre of the remaining spicule solution was mounted onto a slide using DPX and examined at a magnification of ×100–200, using a compound light microscope (Leica Microsystems DM LB) with attached digital camera (Canon EOS 70D). Spicule type (see Supplementary Information for definitions of spicule types) and location (defined as region of tissue in the sponge—cortex versus choanosome) was recorded for a qualitative analysis. For a quantitative comparison, the sizes of 30 of each spicule type which were present in all sponges (strongyloxea, megasters and micrasters) were recorded per sponge. For megascleres (strongyloxea), the length and width were recorded for both thicker and thinner auxiliary types (stronglyoxea and styles, respectively). For both megasters and micrasters, the diameter and R:C ratio (ray length to centre length ratio) were recorded, following Bergquist and Kelly-Borges (1991) and Sàra and Sàra (2004). Size measurements for all megasters (spherasters and oxyspherasters) were grouped, as well as micrasters (oxyasters, chiasters, stronglyasters and tylasters), because they could not be differentiated using light microscopy. Sizes were measured using ImageJ (Schneider et al. 2012). Non-parametric multidimensional scaling (nMDS) ordinations were generated using Euclidean distance on Primer v7 (Clarke 1993) to determine any clustering based on average, minimum and maximum sizes. Spicules from all 18 sponges that were subsampled for spicule analysis were also observed using Scanning Electron Microscopy (SEM), to visualize the megasters and micrasters at higher resolution and magnification. Spicules preserved in ethanol were mounted on stubs, air dried and coated with a gold/palladium alloy using a sputter coater. Scanning electron micrographs were taken with the Hitachi TM3000 Benchtop Scanning Electron Microscope in high vacuum mode.
Results
Molecular phylogeny
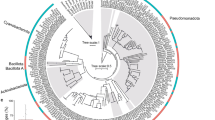
A total of 1420 bp (425 bp for 18S, 570 bp for rnl and 425 bp for COI-ext) were sequenced for 71 Tethya individuals. For all trees, branch confidence (via bootstrap/posterior probability values) ranged from 64 to 100%/0.91 to 1 for divergences between the main clades (Figs. 2 and 3). 18S resolved the clades above 86%/0.96 and rnl above 74%/0.99, but COI-ext exhibited slightly lower confidence from 63%/0.91. The nuclear 18S region grouped sponges into three genotypes, and for the mitochondrial rnl and COI-ext partitions there were seven and four haplotypes present, respectively. The concatenated data set for the mitochondrial markers revealed 15 different haplotypes (Fig. 3). For each individual locus, phylogenetic reconstructions revealed three divergent clades that each contained samples from all sampling locations; in other words, none of the clades contained individuals from strictly one location. For rnl and 18S, those sponges that were thought to be T. bergquistae all grouped together into one clade (Clade 1), but for COI-ext, one individual that was thought to be T. bergquistae grouped with Clade 2 (T. burtoni clade). For T. burtoni, rnl and 18S generated congruent placement of sponges into two separate groups. However, COI-ext placed sponges differently, where some sponges that belonged to Clade 2 for rnl and 18S were placed into Clade 3; and in addition, two sponges which were thought to be T. burtoni were placed into Clade 1 (T. bergquistae clade). The three uncertain yellow sponges (Fig. 1) grouped with T. bergquistae for all three markers.
Phylograms showing the relationship between Tethya bergquistae (Clade 1) and T. burtoni (Clades 2 and 3) based on individual mitochondrial markers (rnl and COI-ext). Topology generated from Maximum Likelihood (ML) analysis. ML bootstrap confidence values/Bayesian Inference (BI) posterior probabilities given on each branch. T. actinia was selected as an outgroup. Inconsistencies in the COI-ext tree versus rnl are highlighted by having sponges retain the colour of the rnl placement and by bolding individuals. Scale bar = substitutions per site
Phylograms showing the relationship between Tethya bergquistae (Clade 1) and T. burtoni (Clades 2A + B and 3) based on nuclear DNA (18S) and mitochondrial DNA (concatenated COI-ext + rnl). Topology generated from Maximum Likelihood (ML) analysis. ML bootstrap confidence values/Bayesian Inference (BI) posterior probabilities given on each branch. T. actinia was selected as an outgroup. Scale bar = substitutions per site
Divergence was calculated between and within all genetic clades using p distances (Table 2). Intraclade p distance across all three markers ranged from 0 to 0.13% (average = 0.04%, stdev = 0.06%), whereas interclade p distances for all pairwise clade comparisons across all markers ranged from 0.5 to 11.2% (average = 5.29%, stdev = 3.83%). For all pairwise clade comparisons, interclade p distances for 18S (0.5–1.2%) were lower than for the mt-markers rnl and COI-ext (4.8–8.6, 6.4–11.2%, respectively). For T. bergquistae (Clade 1), intraclade p distances for all markers was zero. For T. burtoni (Clades 2 + 3, combined), intraclade p distances were: zero for 18S, 2.1% for rnl and 4.2% for COI-ext. The divergence between T. bergquistae and T. burtoni was: 0.8% for 18S, 6.6% for rnl and 9.0% for COI-ext.
For T. burtoni, both PCA and STRUCTURE based on microsatellite allele frequencies revealed two distinct genetic clusters that were congruent with the T. burtoni clades generated from phylogenetic analyses for 18S and rnl (Fig. 4). The T. burtoni clades (Clades 2 and 3) were significantly different with respect to their genetic structure (Fst = 0.39, P < 0.0001). When using only those loci in HWE, differentiation between clades was also significant and strong (Fst = 0.47, P < 0.0001).
Genetic differentiation within Tethya burtoni based on microsatellite data. On left: Principal coordinate analysis (PCA) based on genetic distances from microsatellite allele frequencies for the T. burtoni clades (Clades 2 and 3 from Fig. 3). Each point represents a T. burtoni genotype, and the two groups (C2 and C3) are labelled inside of their 67% inertia ellipses. On right: Group assignment of T. burtoni individuals into four genetic clusters (K = 4) based on STRUCTURE. Each vertical line represents an individual, and the coloration is the proportion of that individual’s estimated membership into each of the four genetic clusters. Labels along the horizontal refer to individual IDs (see Supplementary Table S1 for more information). Horizontal bars under individual IDs correspond to sampling locations, which are: Kapiti Island (black), Red Rocks (dark grey), Breaker Bay (light grey) and Somes Island (white)
Morphological analysis
All sponges contained megascleres that were strongyloxea and anisostronglyes, along with smaller, thinner auxiliary styles. Megasters were spherasters and oxyspherasters, and micrasters were tylasters, chiasters, stronglyasters and oxyasters. Spicule sizes are summarized in Table 3. Average strongyloxea and auxiliary style length × width were 1061 ± 275 × 21 ± 6 µm and 399 ± 116 × 13 ± 4 µm, respectively. Respective megaster and micraster diameters were 44 ± 10 and 14 ± 3 µm, and the average R:C ratio was ~0.8 ± 0.2 for all asters. n-MDS plots for spicule size (average, minimum and maximum sizes) revealed no evident clustering (Fig. 5). Qualitative differences were observed between the location and density of megasters within the different genetic clades. Those sponges belonging to Clade 1 contained megasters throughout both the cortex and choanosome, whereas those sponges in Clades 2 and 3 had megasters in the cortex, but little to none in the choanosome. Within the T. burtoni clades (Clades 2 and 3), there were no differences in spicule composition detectable by light microscopy. Scanning electron micrographs of spicules from the three genetic clades also revealed no visible differences in megastar or micraster morphology (Fig. 6). However, sponges belonging to the T. bergquistae clade contained an additional spined oxyaster, which was absent in the T. burtoni clades. A qualitative assessment of cross sections from this subsample of sponges revealed high plasticity within each genetic clade, and no clear differences between clades (Fig. 1). Further, 20 μm cross sections examining the spicule composition between the choanosome and ectosome revealed that T. bergquistae contained more asters in the choanosome than T. burtoni (Fig. 7). However, within T. burtoni, there were no evident differences between Clades 2 and 3. While some members of Clade 2 appeared to contain a slightly higher density of asters in the cortex, this was not consistent for all individuals within that clade.
Nonparametric-multidimensional (nMDS) scaling ordination of mean spicule sizes for Tethya bergquistae (pink squares, Clade 1) and T. burtoni (orange circles, Clade 2; and blue triangles, Clade 3). Clades are identified in Fig. 3. Spicules sizes are based on measurements from strongyloxea, megasters and micrasters (n = 30 per sponge). Codes above each sample refer to individual sample ID (refer to Supplementary Table S1 for more information on individual samples). A subset of all samples are presented in the nMDS, based on a Euclidean distance matrix
Scanning electron micrographs of spicules belonging to Tethya bergquistae (Clade 1) and T. burtoni (Clade 2 + 3), from sponges belonging to the three genetic clades of Tethya identified in Fig. 3. Clade 1: A–C = spherasters; D = spined oxyaster; E–H = tylasters; G = oxyaster. Clade 2 and 3: A–B = spherasters; C–D = tylasters; E = oxyaster. Scale bars are located in the bottom right of each micrograph and represent 10 µm
Discussion
Here, we document discordance between molecular and morphological descriptions of Tethya spp. in central New Zealand and highlight the potential for cryptic speciation to go undetected based on morphological features alone. Phylogenetic reconstructions based on 18S, rnl and COI-ext revealed three genetic clades, with a speciation event likely occurring in the group described as T. burtoni. This division within T. burtoni was further confirmed using novel microsatellite markers. Morphological analyses based on spicule size and composition allowed T. bergquistae to be differentiated from T. burtoni but failed to reveal differences between the T. burtoni clades. To our knowledge, this is only the second report of cryptic species on the basis of molecular differences within the group Tethya (with the first being T. robusta, Sarà et al. 1993). Understanding how organisms evolve is central to ecology and evolution, but can be particularly complex for phenotypically similar organisms living in sympatry with no obvious barriers to reproduction, reflected in Tethya spp. examined here. Overall, our study highlights the need to use more than one method to define sponge species boundaries in a reliable way.
Species delineation based on genetics
The phylogenetic relationship of Tethya spp. was not congruent for all three markers, as COI-ext placed sponges into different clades with most discrepancies occurring in the T. burtoni group. Microsatellite markers also supported the T. burtoni clade division that was indicated in the 18S and rnl trees (Figs. 2 and 3). The use of COI to delineate species has had differential success among sponges (Erpenbeck et al. 2006b; Wörheide and Erpenbeck 2007; Erpenbeck et al. 2007; Poppe et al. 2010; Belinky et al. 2012) as well as other organisms (Shearer and Coffroth 2008; Derycke et al. 2010), resulting in debate around its use as an informative marker (Waugh 2007). The COI region often has low variability (Bucklin et al. 2011), and this conservativeness can fail to capture evolutionary relationships between closely related species. We instead employed COI-ext to avoid this drawback, as this region downstream of COI is thought to substitute earlier during species divergence (Erpenbeck et al. 2006a). We captured variation between T. bergquistae and T. burtoni, as well as within T. burtoni; however, the relationship produced from COI-ext was incongruent with that produced from 18S, rnl and the 11 microsatellite markers. For species living in sympatry, introgression can be common, resulting in semipermeable species boundaries (Rüber et al. 2001, Harrison and Larson 2014). Genes can introgress at different rates (called differential introgression), which can result in phylogenies from multiple markers in disagreement (Harrison and Larson 2014). It is possible that the inconsistency in the COI-ext phylogeny is a product of this, and if true, gives evidence of introgression between T. bergquistae and T. burtoni. For instance, one sponge (KP72) that was thought to be T. bergquistae (as evidenced by its placement into Clade 1 for 18S and rnl) was placed into the T. burtoni clade (Clade 2) for COI-ext. In addition, two sponges (BB177 and SI322), which were thought to be T. burtoni (from their placement into Clade 2 by 18S and rnl), were placed into the T. bergquistae clade (Clade 1) for COI-ext. Within T. burtoni, there is more disagreement between COI-ext and the other markers within the clades. For example, those sponges belonging to Clade 2B of the mitochondrial concatenated data set (Fig. 3) were placed into Clade 2 by 18S, rnl and the microsatellite markers, but Clade 3 for COI-ext. It is possible that this group of sponges, which possess genes common to two different putative species (discussed below), have also undergone introgressive hybridization. Some corals have also been found to exhibit high degrees of sympatry, cryptic speciation and introgression (Ladner and Palumbi 2012), and perhaps shared life-history traits between corals and sponges (i.e. sessile, sexual and asexual reproductive ecology) shape the evolutionary history of these populations. While additional sequencing work would provide clarity on this topic, it appears highly probable that the species boundaries between T. bergquistae and T. burtoni, as well as between the putative T. burtoni cryptic species, are actually semipermeable.
The definition of a species continues to be debated, and when complex evolutionary processes like introgression can shape species, understanding species delineations becomes more complicated. The topology produced from 18S clearly differentiates our clades. 18S is a slowly evolving gene region (Berntson et al. 2001), and as such often captures interspecific variation rather than intraspecific variation. Three separate clades, despite the slow evolving nature of this gene, provides strong evidence that these three clades correspond to three different species. Most interestingly, two clades exist within T. burtoni and suggest cryptic speciation within the group. Other measures exist (e.g. p distances, Fst) to more definitively set a quantitative value to define species divisions. Comparing the p distances obtained in this study to other observed divergence values for sponges (Table 4), both within- and between-clade distances give further evidence for species-level relatedness. Our intraclade p distances were about ~5–50 times less than the interclade values for all pairwise comparisons of clades, which is consistent with intraspecific values for other sponge taxa. These intraclade distances suggest that sponges belonging to the same clade are conspecific. When comparing the individual markers used here to the same markers (or similarly evolving markers) in other studies, the interclade p distances calculated for all pairwise clade comparisons in this study compare to interclade p distances for other sponge species (Table 4). This further reveals our clades do in fact correspond to species. For example, Scopalina lophyropoda was reported as being comprised of a cryptic species complex (Blanquer and Uriz 2007), and the interclade p distances for 18S (0.3–2%) were on the same scale as for those clades we found within T. burtoni (0.5%). Considering T. bergquistae (Clade 1) versus T. burtoni (Clades 2 + 3, combined), divergence between the two species was: 0.8% for 18S, 6.6% for rnl and 9% for COI-ext. Divergence between the T. burtoni clades (Clade 2 versus Clade 3) were on the same order: 0.5% for 18S, 4.8% for rnl and 8.3% for COI-ext. Divergence within T. burtoni that is on the same scale as between T. burtoni and T. bergquistae gives further evidence that the two T. burtoni clades correspond to different species. The microsatellite data also reveal strong differentiation within the T. burtoni group (Fst = 0.39, P < 0001). While microsatellite markers are traditionally not used as a tool for phylogenetic studies (but instead population structure analysis), used here in combination with the other genetic markers gives further evidence for strong genetic differentiation between clades. Interestingly, differentiation between both T. burtoni clades was not a product of location (population structure of T. burtoni discussed elsewhere), as evident from the STRUCTURE plot (Fig. 4). Instead, sponges that lived sympatrically were strongly genetically different, suggesting reproductive isolation for sponges in the same location. Cryptic speciation has been recorded throughout the Porifera phylum (Solé-Cava et al. 1991; Muricy et al. 1996; Nichols and Barnes 2005; Wörheide et al. 2008; Xavier et al. 2010; DeBiasse and Hellberg 2015; Knapp et al. 2015; Uriz et al. 2017), and is also likely for what’s known as T. burtoni. We believe our genetic data combined gives strong evidence that T. burtoni is in fact be comprised of a species complex, being more genetically diverse than previously thought based on morphology alone.
Discordance between molecular and morphological characteristics
Our analysis of spicule size/type, in conjunction with our qualitative assessment of macromorphological features (colour and cross sections), was inconsistent with the molecular phylogeny that we generated. Macromorphological features allowed sponges belonging to the T. bergquistae clade to be distinguished from sponges belonging to the T. burtoni clades; however, we were not able to identify sponges belonging to separate genetic clades within T. burtoni from morphology. The three uncertain, bright yellow specimens grouped together with those sponges belonging to T. bergquistae, which has not previously been described as yellow (Bergquist and Kelly-Borges 1991; Hooper and Wiedenmayer 1994; Battershill et al. 2010), indicating that colour is not a reliable distinguishing feature in this case. Qualitative observations revealed no obvious differences in cross sections between T. burtoni clades, and instead were highly variable within each clade. While generally cross sections are a key defining feature between species of Tethya, they did not provide clear resolution here. There was a marked difference between the spicule composition of T. bergquistae and the T. burtoni complex (more asters in the choanosome of the former), but there were no differences evident within the T. burtoni clades. Morphological characteristics therefore provided no clear evidence of speciation within T. burtoni. Because there is a lack of defining characteristics available for Porifera, it is common for genetic diversity (and cryptic speciation) to go undetected, where molecular and morphological relationships are commonly incongruent (McCormack et al. 2002; Erpenbeck et al. 2006b; Xavier et al. 2010). Determining which morphological traits are informative in species differentiation can also be complicated. For example, Erpenbeck et al. (2006b) documented that sponges containing highly divergent skeletal features are actually closely genetically related. In contrast, spicules within the same species may be highly variable due to environmental influences (Bell et al. 2002). We show here that morphological features may not be able to reveal species boundaries, as perhaps skeletal changes following divergence may be delayed. Mutations may accumulate without altering the phenotype when nucleotide changes have no effect on how its product (protein) folds and its function overall (Avise 2012), which could explain delayed morphological expression in genetically different individuals. While it is possible that subtle morphological differences may exist, they were not able to be detected in this study. The high phenotypic plasticity observed across individuals of T. burtoni resulted in a lack of clear diagnostic features for sponges belonging the different genetic clades. Furthermore, the potential introgression events between cryptic species may make detecting morphological differences between such species more difficult. To avoid this potential confounding factor, we used sponges which showed no introgression based on our markers for the morphological analysis presented here. However, we cannot confirm that these sponges have not undergone introgression, as we only surveyed three genes. A survey of other regions of the genome may reveal clearer relationships and show the scope of introgressive hybridization between the T. burtoni cryptic species. This may shed more light on differences between plasticity versus distinct morphological features that allow species to be characterized. With such incongruence between morphological and molecular data, it is imperative to use a combination of methods to correctly delineate species in order to fully understand the species evolutionary relationships.
Conclusions
Our study highlights the complexity in delineating sympatric, morphologically similar species when disagreement between morphological and molecular traits exist, and the importance of using multiple taxonomic methods. While traditional morphology-based taxonomy can be the first step in species identification, we show that it can fail to reveal cryptic species. Being able to confidently identify a species is crucial to any ecological or evolutionary study, yet this remains a challenge. Organisms with few defining traits, like sponges, present an opportunity to examine potential morphological and molecular discord, and highlight the complexity in defining species boundaries. Failure to detect cryptic species may result in an underestimation of biodiversity and an incorrect interpretation of functional roles of an organism. In addition, it can mislead our understanding of connectivity patterns and evolutionary relationships, if interspecific diversity is confused for intraspecific variation. As such, we strongly recommend the use of integrative taxonomy in species identification.
References
Adlard RD, Lester JG (1995) Development of a diagnostic test for Mikrocytos roughleyi, the aetiological agent of Australian winter mortality of the commercial rock oyster, Saccostrea commercialis (Iredale & Roughley). J Fish Dis 18:609–614
Alvarez B, Crisp MD, Driver F, Hooper JNA, Van Soest RWM (2000) Phylogenetic relationships of the family Axinellidae (Porifera: Demospongiae) using morphological and molecular data. Zool Scr 29:169–198
Andreakis N, Luter HM, Webster NS (2012) Cryptic speciation and phylogeographic relationships in the elephant ear sponge Ianthella basta (Porifera, Ianthellidae) from northern Australia. Zool J Linn Soc 166:225–235
Avise JC (2012) Molecular markers, natural history and evolution. Springer Science & Business Media, New York, NY
Battershill CN, Bergquist PR, Cook SDC (2010) Phylum Porifera. In: Cook SDC (ed) New Zealand coastal marine invertebrates, vol 1. Canterbury University Press, Christchurch, New Zealand, pp 122–124.
Baum DA, Shaw KL (1995) Genealogical pers pectives on the species problem. In: Hoch PC, Stevenson AG (eds) Experimental and molecular approaches to plant biosystematics, vol 53. Missouri Botanical Garden, St. Louis, pp 289–303
Bavestrello G, Bonito M, Sará M (1993) Influence on depth on the size of sponge spicules. Sci Mar 57:415–420
Belinky F, Szitenberg A, Goldfarb I, Feldstein T, Wörheide G, Ilan M et al. (2012) ALG11 – a new variable DNA marker for sponge phylogeny: Comparison of phylogenetic performances with 18S rDNA and the COI gene. Mol Phylogenet Evol 63:702–713
Bell J, Barnes D, Turner J (2002) The importance of micro and macro morphological variation in the adaptation of a sublittoral demosponge to current extremes. Mar Biol 140:75–81
Bergquist PR (1961) Demospongiae (Porifera) of the Chatham Islands and Chatham Rise, collected by the Chatham Islands 1954 Expedition. NZ Oceanogr Inst Mem 13:169–206
Bergquist PR, Kelly-Borges M (1991) An evaluation of the genus Tethya (Porifera: Demospongiae: Hadromerida) with descriptions of new species from the southwest Pacific. Beagle, Rec North Territ Mus Arts Sci 8:37–72
Berntson EA, Bayer FM, McArthur AG, France SC (2001) Phylogenetic relationships within the Octocorallia (Cnidaria: Anthozoa) based on nuclear 18S rRNA sequences. Mar Biol 138:235–246
Bickford D, Lohman DJ, Sodhi NS, Ng PL, Meier R, Winker K et al. (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22:148–155
Blanquer A, Uriz MJ (2007) Cryptic speciation in marine sponges evidenced by mitochondrial and nuclear genes: a phylogenetic approach. Mol Phylogenetics Evol 45:392–397
Bond C (1992) Continuous cell movements rearrange anatomical structures in intact sponges. J Exp Zool A Ecol Integr Physiol 263:284–302
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27:1767–1780
Bucklin A, Steinke D, Blanco-Bercial L (2011) DNA barcoding of marine metazoan. Ann Rev Mar Sci 3:471–508
Capa M, Pons J, Hutchings P (2013) Cryptic diversity, intraspecific phonetic plasticity and recent geographical translocations in Branchiomma (Sabellidae, Annelida). Zool Scr 42:637–655
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Corriero G, Gadaleta F, Bavestrello G (2015) A new Mediterranean species of Tethya (Porifera: Tethyida: Demospongiae). Ital J Zool 82:535–543
Cruz-Barraza JA, Carballo JL, Rocha-Olivares A, Ehrlich H, Hog M (2012) Integrative taxonomy and molecular phylogeny of genus Aplysina (Demospongiae: Verongida) from Mexican Pacific PLoS ONE 7:e42049
Czarna A, Gawrońska B, Nowińska R, Morozowska M, Kosiński P (2016) Morphological and molecular variation in selected species of Erysimum (Brassicaceae) from Central Europe and their taxonomic significance. Flora 222:68–85
De Paula TS, Zilberberg C, Haidu E, Lobo-Haidu G (2012) Morphology and molecules on opposite sides of the diversity gradient: four cryptic species of the Cliona celata (Porifera, Demospongiae) complex in South America revealed by mitochondrial and nuclear markers. Mol Phylogenet Evol 62:529–541
De Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56:879–886
DeBiasse MB, Hellberg ME (2015) Discordance between morphological and molecular species boundaries among Caribbean species of the reef sponge Callyspongia. Ecol Evol 5:663–675
Derycke S, Vanaverbeke J, Rigaux A, Backeljau T, Moens T (2010) Exploring the use of cytochrome oxidase c subnit 1 (COI) for DNA barcoding of free-living marine nematodes. PLoS ONE 5:e13716. https://doi.org/10.1371/journal.pone.0013716
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Erpenbeck D, Hooper JNA, Wörheide G (2006a) CO1 phylogenies in diploblasts and the ‘Barcoding of Life’— are we sequencing a suboptimal partition? Mol Ecol Notes 6:550–553.
Erpenbeck D, Breeuwer JAJ, Parra-Velandia FJ, Van Soest RWM (2006b) Speculation with speculation? Three independent gene fragments and biochemical characters versus morphology in demosponge higher classification. Mol Phylogenet Evol 38:293–205.
Erpenbeck D, Duran S, Rutzler K, Paul V, Hooper JNA, Wörheide G (2007) Towards a DNA taxonomy of Caribbean demosponges: a gene tree reconstructed from partial mitochondrial CO1 gene sequences supports previous rDNA phylogenies and provides a new perspective on the systematics of Demospongiae. J Mar Biol Assoc UK 87:1563–1570
Erwin PM, Thacker RW (2007) Phylogenetic analyses of marine sponges within the order Verongida: a comparison of morphological and molecular data. Invertebr Biol 126:220–234
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Frankham R, Ballou JD, Dudash MR, Eldridge MDB, Fenster CB, Lacy RC et al. (2012) Implications of different species concepts for conserving biodiversity. Biol Conserv 153:25–31
Gómez A, Serra M, Carvalho GR, Lunt DH (2002) Speciation in ancient cryptic species complexes: evidence from the molecular phylogeny of Branchionus plicatilis (Rotifera). Evolution 56:1431–1444
Harrison RG, Larson EL (2014) Hybridization, introgression and the nature of species boundaries. J Hered 105:795–809
Hebet PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA 41:14812–14817
Heim I, Nickel M, Brümmer F (2007) Phylogeny of the genus Tethya (Tethyidae: Hadromerida: Porifera): molecular and morphological aspects. J Mar Biol Assoc UK 87:1615–1627
Hennig W (1966) Phylogenetic systematics. University of Illinois Press, Urbana
Hey J, Waples RS, Arnold ML, Butlin RK, Harrison RG (2003) Understanding and confronting species uncertainty in biology and conservation. Trends Ecol Evol 18:597–603
Hilis DM (1987) Molecular versus morphological approaches to systematics. Annu Rev Ecol Evol Syst 18:23–42
Hooper JNA (2003) ‘Sponguide’. A guide to sponge collection and identification. In: Hooper JNA, Van Soest RWM (eds) 2002. Systema Porifera. A guide to the classification of sponges, vol 1–2. Kluwer Academic/Plenum Publishers, New York, NY, Boston, Dordrecht, London, Moscow, pp xlviii–1708.
Hooper JNA, Wiedenmayer F (1994) Porifera. In: Wells A (ed) Zoological catalogue of Australia, vol 12. CSIRO, Melbourne, pp 1–620.
Hooper JNA, Van Soest RWM (2002) Systema Porifera. a guide to the classification of sponges. Kluwer Academic/Plenum Publishers, New York, NY, Boston, Dordrecht, London, Moscow
Hulce D, Li X, Snyder-Leiby T (2011) GeneMarker genotyping software: tools to increase the statistical power of DNA fragment analysis. J Biomol Tech 22:S35–S36
Jakobsson M, Rosenberg NA (2007) CLUMPP: a clustering matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T (2008) Adegenet: R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Kauserud H, Svegarden IB, Decock C, Hallenberg N (2006) Hybridization among cryptic species of the cellar fungus Coniophora puteana (Basidiomycota). Mol Ecol 16:389–399
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Knapp IS, Forsman ZH, Williams GJ, Toonen RJ, Bell JJ (2015) Cryptic species obscure introduction pathway of the blue Carribean sponge (Haliclona (Soestella) caerulea), (order: Haplosclerida) to Palmyra- Atoll, Central Pacific. Peer J 3:e1170
Knowlton N (1993) Sibling species in the sea. Annu Rev Ecol Syst 24:189–216
Knowlton N (2000) Molecular genetic analyses of species boundaries in the sea. Hydrobiologia 420:73–90
Kumar S, Stecher G, Tamura K (2015) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Ladner JT, Palumbi SR (2012) Extensive sympatry, cryptic diversity and introgression throughout the geographic distribution of two coral species complexes. Mol Ecol 21:2224–2238
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. (2007) ClustalW and ClustalX. Bioinformatics 23:2947–2948
Lavrov DV, Wang XJ, Kelly M (2008) Reconstructing ordinal relationships in the Demospongiae using mitochondrial genomic data. Mol Phylogenet Evol 49:111–124
Lecompte E, Denys C, Granjon L (2005) Confrontation of morphological and molecular data: the Praomys group (Rodentia, Murinae) as a case of adaptive convergences and morphological stasis. Mol Phylogenet Evol 37:899–919
Ledenfield R (1888) Descriptive catalogue of sponges in the Australian Museum, Sydney. Taylor and Francis, London
Lihová J, Kučera J, Perný M, Marhold K (2007) Hybridization between two polypoloid Cardamine (Brassicacea) species in north-western Spain: Discordance between morphological and genetic variation patterns. Ann Bot 99:1083–1096
Mayden RL (1997) A hierarchy of species concepts: the denouement in the saga of the species problem. In: Claridge MF, Dawah HA, Wilson MR (eds) Species: the units of biodiversity. Chapman and Hall, London, pp 381–424.
Mayr E (1942) Systematics and the origin of species. Columbia University Press, New York, NY
McCormack GP, Erpenbeck D, Van Soest RWM (2002) Major discrepancy between phylogenetic hypotheses based on molecular and morphological criteria within the Order Haplosclerida (Phylum Porfera: Class Demospongiae). J Zool Syst Evol Res 40:237–240
McDonald JI, Hooper JNA, McGuinness KA (2002) Environmentally influenced variability in the morphology of Cinachyrella australiensis (Carter 1886) (Porifera: Spirophorida: Tellidae). Mar Freshw Res 53:79–84
Morrow C, Cárdenas P (2015) Proposal for a revised classification of the Demospongiae (Porifera). Front Zool 12:7
Muricy G, Solé-Cava AM, Thorpe JP, Boury-Esnault N (1996) Genetic evidence for extensive cryptic speciation in the subtidal sponge Plakina trilopha (Porifera: Demospongiae: Homoscleromorpha) from the Western Mediterranean. Mar Ecol Prog Ser 138:181–187
Nakayama S, Arima K, Kawai K, Mohri K, Inui C, Sugano W et al. (2015) Dynamic transport and cementation of skeletal elements build up the pole-and-beam structured skeleton of sponges. Curr Biol 25:2549–2554
Nichols SA, Barnes PAG (2005) A molecular phylogeny and historical biogeography of the marine sponge genus Placospongia (Phylum Porifera) indicate low dispersal capabilities and widespread crypsis. J Exp Mar Biol Ecol 323:1–15
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University: Sweden.
Padial JM, Miralles A, De la Rive I, Vences M (2010) The integrative future of taxonomy. Front Zool 7:16
Poppe J, Sutcliffe P, Hooper JNA, Wörheide G, Erpenbeck D (2010) COI barcoding reveals new clades and radiation patterns of Indo-Pacific sponges of the family Irciniidae (Demospongiae: Dictyoceratida). PLoS ONE 5:e9950
Plotkin A, Voight O, Willassen E, Rapp HT (2017) Molecular phylogenies challenge the classification of Polymastiidae (Porifera, Demospongiae) based on morphology. Org Divers Evol 17:45–66
Pritchard K, Ward V, Battershill C, Bergquist PR (1984) Marine sponges: forty-six sponges of Northern New Zealand. Leigh Lab Bull 14:149
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Redmond NE, McCormack GP (2009) Ribosomal internal transcriber spacer regions are not suitable for intra- or inter-specific phylogeny reconstruction in haplosclerid sponges (Porifera: Demospongiae). J Mar Biol Assoc UK 89:1251–1256
Reveillaud J, Remerie T, Van Soest R, Erpenbeck D, Cardenas P, Derycke S et al. (2010) Species boundaries and phylogenetic relationships between Altanto-Mediterranean shallow-water and deep-sea coral associated with Hexadella species (Porifera, Ianthellidae). Mol Phylogenet Evol 56:104–114
Reveillaud J, van Soest R, Derycke S, Picton B, Rigaux A, Vanreusel A (2011) Phylogenetic relationships among NE Atlantic Plocamionida Topsent (1927) (Porifera, Poecilosclerida): under-estimated diversity in reef ecosystems. PLoS ONE 6:e16533
Ronquist F, Teslenko M, Van der Mark P, Ayres D, Darling A, Höhna S et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Rosen DE (1979) Fishes from the uplands and intermontane basins of Guatemala: revisionary studies and comparative geography. Bull Am Mus Nat Hist 162:267–376
Rot C, Goldfarb I, Ilan M, Huchon D (2006) Putative cross-kingdom horizontal gene transfer in sponge (Porifera) mitochondria. BMC Evol Biol 6:71
Rüber L, Meyer A, Sturmbauer C, Verheyen E (2001) Population structure in two sympatric species of the Lake Tanganyika cichlid tribe Eretmodini: evidence for introgression. Mol Ecol 10:1207–1225
Sarà M (1987) A study on the genus Tethya (Porifera Demospongiae) and new perspectives in sponge systematics. In: Vacelet J, Boury-Esnault N (eds) Taxonomy of Porifera. NATO ASI Series (Series G: Ecological Sciences), vol 13. Springer, Berlin.
Sarà M (1998) A biogeographic and evolutionary survey of the genus Tethya (Porifera, Demospongiae). In: Watanabe Y, Fusetani N (eds) Sponge sciences, vol 1. Springer, Tokyo, pp 83–94.
Sarà M, Corriero G, Bavestrello G (1993) Tethya (Porifera, Demospongiae) species coexisting in a Maldivian coral reef lagoon: Taxonomical, genetic and ecological data. Mar Ecol 14:341–355
Sarà M, Sarà A (2004) A revision of Australian and New Zealand Tethya (Porifera: Demospongiae) with preliminary analysis of species-groupings. Invertebr Syst 18:117–156
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Shearer TL, Coffroth MA (2008) DNA barcoding: barcoding corals: limited by interspecific divergence, not intraspecific variation. Mol Ecol Res 8:247–255
Simpson GG (1951) The species concept. Evolution 5:285–298
Solé-Cava AM, Klautau M, Boury-Esnault N, Borojecic R, Thorpe JP (1991) Genetic evidence for cryptic speciation in allopatric populations of two cosmopolitan species of the calcareous sponge genus Clathrina. Mar Biol 111:381–386
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinaeur Associates, Sunderland, Massachusetts
Uriz MJ, Garate L, Agell G (2017) Molecular phylogenies confirm the presence of two cryptic Hemimycale species in the Mediterranean and reveal the polyphyly of the genera Crella and Hemimycale (Demospongiar: Poecilosclerida). Peer J 5:e2958
Van Soest RWM, Boury-Esnault N, Hooper JNA, Rützler K, de Voogd NJ, Alvarez B, et al. (2018) World Porifera database. Accessed at http://www.marinespecies.org/porifera/porifera.php?p=taxdetails&id=132077 on 2017-10-14
Van Valen L (1976) Ecological species, multispecies, and oaks. Taxon 25:233–239
Vrijenhoek RC (2009) Cryptic species, phenotypic plasticity, and complex life histories: assessing deep-sea faunal diversity with molecular markers. Deep-Sea Res Part II 56:1713–1723
Wallis GP, Cameron‐Christie SR, Kennedy HL, Palmer G, Sanders TR, Winter DJ (2017) Interspecific hybridization causes long‐term phylogenetic discordance between nuclear and mitochondrial genomes in freshwater fishes. Mol Ecol 26:3116–3127
Waples RS (2015) Testing for Hardy-Weinberg proportions: have we lost the plot? J Hered 106:1–19
Waugh J (2007) DNA barcoding in animal species: progress, potential and pitfalls. Phylogenet Syst 29:188–197
Wendel JF, Doyle JJ (1998) Phylogenetic incongruence: window into genome history and molecular evolution. In: Soltis DE, Soltis PS, Doyle JJ (eds) Molecular systematic of plants II. Springer, Boston
Wörheide G, Erpenbeck D (2007) DNA taxonomy of sponges – progress and perspectives. J Mar Biol Assoc UK 87:1629–1633
Wörheide G, Epp LS, Macis L (2008). Deep genetic divergence among Indo-Pacific populations of the coral reef sponge Leucetta chagoensis (Leucettidae): founder effects, vicariance, or both? BMC Evol Biol 8: https://doi.org/10.1186/1471-2148-8-24.
Xavier JR, Rachello-Dolmen PG, Parra-Velandia F, Schönberg CHL, Breeuwer JAJ, van Soest RWM (2010) Molecular evidence of cryptic speciation in the “cosmopolitan” excavating sponge Cliona celata (Porifera, Clionaidae). Mol Phylogenet Evol 56:13–20
Wörheide G (2006) Low variation in partial cytochrome oxidase subunit I (COI) mitochondrial sequences in the coralline demosponge Astrosclera willeyana across the Indo-Pacific. Mar Biol 148:907–912
Acknowledgements
This research was funded by a Victoria University of Wellington Doctoral Scholarship awarded to Megan Shaffer. A special thanks to Dr. Michael Gardner and Tessa Bradford (Flinders University) for their help with microsatellite development, and to Daniel Leduc (National Institute of Water and Atmospheric Research) for training and use of the SEM. We are also grateful to Dr. Manuel Maldonado (Center for Advanced Studies of Blanes, CEAB-CSIC), who offered insight into the interpretation of our results. Lastly, thanks to the technicians at the Victoria University Coastal Ecology Lab (Daniel McNaughtan, John van der Sman, Dan Crossett) and to Bucky Focht, who aided in the collection of these samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability
Sequence data have been submitted to GenBank, under the accession numbers MH180010-180023. Microsatellite genotype data is available on Pangaea (https://doi.org/10.1594/PANGAEA.889125).
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Shaffer, M.R., Davy, S.K. & Bell, J.J. Hidden diversity in the genus Tethya: comparing molecular and morphological techniques for species identification. Heredity 122, 354–369 (2019). https://doi.org/10.1038/s41437-018-0134-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-018-0134-6