Abstract
One of the most economically important areas within the Russian agricultural sector is dairy and beef cattle farming contributing about $11 billion to the Russian economy annually. Trade connections, selection and breeding have resulted in the establishment of a number of breeds that are presumably adapted to local climatic conditions. Little however is known about the ancestry and history of Russian native cattle. To address this question, we genotyped 274 individuals from 18 breeds bred in Russia and compared them to 135 additional breeds from around the world that had been genotyped previously. Our results suggest a shared ancestry between most of the Russian cattle and European taurine breeds, apart from a few breeds that shared ancestry with the Asian taurines. The Yakut cattle, belonging to the latter group, was found to be the most diverged breed in the whole combined dataset according to structure results. Haplotype sharing further suggests that the Russian cattle can be divided into four major clusters reflecting ancestral relations with other breeds. Herein, we therefore shed light on to the history of Russian cattle and identified closely related breeds to those from Russia. Our results will facilitate future research on detecting signatures of selection in cattle genomes and eventually inform future genetics-assisted livestock breeding programs in Russia and in other countries.
Similar content being viewed by others
Introduction
Thousands of years of artificial selection coupled with human-driven migration and adaptation to diverse environmental conditions resulted in ~1000 cattle breeds worldwide, which are tailored to local economic needs, aesthetic demands and possess unique genetic profiles (Mason 1969). During the last two centuries, some cattle populations were further improved resulting in several commercial breeds demonstrating outstanding productivity when properly handled (Boichard and Brochard 2012). Currently there is a tendency to replace or ‘improve’ local breeds with the genetic material from superior commercial ones, meaning that genetic diversity, signatures of adaptations to local conditions, and the history of formation encoded in native breed genomes often diminish before being recorded and properly studied (Gaouar et al. 2015). On the other hand, genomes of native breeds could be mined for combinations of the genetic variants invaluable in the development of a new generation of commercial breeds that would better fit into a range of environmental conditions (Gao et al. 2017). The first step towards uncovering this information is to understand the origin, structure and admixture events involving the native breed populations and to place them into the context of a wider set of world breeds (Bovine HapMap Consortium 2009; Matukumalli et al. 2009; Beynon et al. 2015).
The genetic diversity of domestic cattle stems from the two main sources of domestication of the ancient Bos subspecies: B. taurus and B. indicus originating from the Fertile Crescent and the Indus Valley, respectively, and adapted to distinct environments (Loftus et al. 1994). Some extant breeds originate from old and/or recent interbreeding between the B. taurus and B. indicus resulting in a wide geo-climatic adaptation of the hybrids (Larkin and Yudin 2016).
According to a recent study involving the whole-genome genotyping of 129 bovine breeds (Decker et al. 2014), the European cattle breed pool consists mainly of animals of B. taurus ancestry without a great deal of contribution from B. indicus genes, with the exception of Turkish breeds. In addition, the Iberian populations of cattle also have a significant genetic component tracing back to the African taurines (Decker et al. 2014). This comprehensive study, however, did not include breeds from Russia, despite some of them expressing unique adaptations (e.g., the ability to live above the polar circle expressed by the Yakut cattle). Other recent studies of native European cattle did however include a limited number of samples from several Russian native breeds (Iso-Touru et al. 2016; Zinovieva et al. 2016; Upadhyay et al. 2017) but did not carry out a comprehensive comparison between the Russian cattle and the world breeds. A high divergence of the Yakut cattle (Iso-Touru et al. 2016) was suggested as well as distinct genetic profiles of several Russian breeds placing some of them apart from the European Holstein-Friesian population (Zinovieva et al. 2016).
Due to Russia’s unique geographic position in both Europe and Asia, its large territory, diverse climate conditions and its rich history, it is expected that Russian native cattle will demonstrate a variety of adaptations and are likely to form a link between the European and Asian cattle populations. According to historical records, the extant Russian cattle breeds originate from the ancient Eurasian cattle, including the steppe cattle (Li and Kantanen 2010) and later (starting from the early 18th century) were affected by ‘uncontrolled’ interbreeding with multiple European cattle populations (Dmitriev and Ernst 1989). Currently there are 16 native breeds recognised in Russia (Dunin and Dankvert 2013) with even more being extinct (DAD-IS 2017). The Russian cattle breeds can be classified as the breeds of Eastern European origin (e.g., Kholmogory and Yaroslavl), crossbred Eastern European breeds (e.g., Istoben and Kazakh Whiteheaded), and Asian/Siberian/Turano-Mongolian breeds (e.g., Yakut and Buryat) (Buchanan and Lenstra 2015). A comprehensive molecular genetic study of the Russian cattle is missing or limited to the studies based on mitochondrial DNA and a small number of autosomal (Li and Kantanen 2010), and Y-chromosome microsatellite markers (Edwards et al. 2011).
The aim of this study therefore was to analyse a dataset composed mostly of Russian and native breeds from neighbouring countries in the context of the dataset of world breeds. We used the GGP HD150K and Illumina Bovine 50K arrays to genotype individuals from 18 breeds bred in Russia, combined our data with the dataset containing additional 129 cattle breeds collected from around the world (Decker et al. 2014) and samples from ten breeds from Russia and Europe genotyped previously (Iso-Touru et al. 2016). We aimed at building on these established resources to use them as a reference to reveal the genetic structure and history of Russian native cattle and to develop hypotheses about their relationships with breeds worldwide. To reveal the complex history of Russian cattle breeds, multiple complementary methods of population genetics were applied to the datasets, and hypotheses pertaining to the origin and structure of the extant breeds were built based on integration of the results.
Materials and methods
Sample collection
We used breed society and herdbook information to locate the herds of nine native cattle breeds bred in Russia and the Siberian population of Herefords. Collection of blood (maximum volume = 10 ml) was carried out by superficial venepuncture using sterile 10-ml BDK2EDTA Vacutainers® (Wellkang Ltd, London, UK). In addition, sperm samples from bulls of seven breeds were purchased from breeding companies, and sperm samples from six breeds were obtained from Russian Research Institute of Farm Animal Genetics and Breeding (St. Petersburg, Russia). Additional DNA samples for three breeds were identified from the Russian Cattle Genomic Diversity Panel v.1.0 (Yudin et al. 2015). Where pedigree details were available, we attempted to avoid sampling of individuals known to be closely related (e.g., siblings, parent and offspring). Additionally, a balanced combination of the same breed samples from different sources/locations was selected for genotyping (Table 1), however for seven breeds the number of samples collected was <10 with as few as two for the Red Pied cattle suggesting that sampling may not account completely for the breed’s genetics. Whole blood and sperm were both stored at −80 °C until further use.
DNA extraction and genotyping of single nucleotide polymorphisms (SNPs)
DNA from blood samples was extracted using cell lysation followed by phenol-chloroform extraction (Sambrook et al. 2006). The semen samples were pretreated with guanidinium thiocyanate (AppliChem, Darmstadt, Germany) and DNA was extracted using a salting out method (Miller et al. 1988). DNA quality and quantity were determined using a NanoDrop 2000c (Thermo Scientific, Wilmington, DE, USA). High quality samples (i.e., having DNA concentrations of at least 50 ng/µl and A260/280 ratios of ca. 1.8) were then subjected to array genotyping. When the number of DNA samples from purebred unrelated animals of the same or different source/location (Table 1) exceeded ten for a breed, genotyping was performed on the GeneSeek Genomic Profiler High-Density (GGP HD150K) array containing ~139,000 SNP markers with plans to include this dataset into the follow-up studies on detecting signatures of selection in bovine genomes which would benefit from a higher number/density of SNPs being genotyped. Otherwise, samples were genotyped on the BovineSNP50 Analysis BeadChip (BovineSNP50K) array containing ~54,000 SNP markers compatible with many previously published datasets, which is a number sufficient for the present study. Each genotyping set contained several duplicated DNA samples (three for GGP HD150K and two for BovineSNP50K) to control for the quality of genotyping and to identify potentially problematic SNP markers.
Genotypes were called using the GenomeStudio 2 software (Illumina, San Diego, USA), and samples with call rates of <95% were excluded from the further analyses. A pedigree (.ped) file containing the genotype calls, sample and family identifiers and a map (.map) file containing the chromosomal location and identifier for each SNP were generated using GenomeStudio 2 and imported into the PLINK whole genome analysis toolkit (Purcell et al. 2007) for further processing.
Data merging and filtering
To identify relationships between the Russian cattle breeds and worldwide breed collections our GGP HD150K and BovineSNP50K genotyping sets were combined with a set of 48 samples originating from the Ukrainian Grey cattle (Boussaha et al. 2015) applying the PLINK --merge command and a common set of ~43,000 SNP markers shared between the GGP HD150K and BovineSNP50K arrays. To the merged set we added the genotyping sets generated by Decker et al. (2014) (128 additional breeds) and Eurasian breeds from Iso-Touru et al. (2016) (10 breeds). The latter two datasets contained a total of 1836 individuals. The datasets were combined with the PLINK --merge command using only SNP with unique IDs and chromosomal positions as identified by the SNPchiMp v.3 software (Nicolazzi et al. 2015) and custom Python scripts. The combined dataset was further filtered to exclude duplicate samples, poorly genotyped individuals (<95% of SNPs), loci genotyped in <99% of individuals and rare alleles (MAF < 0.001) in PLINK --geno 0.01 --mind 0.05 --maf 0.001 resulting in a subset of 26,740 SNP that were used for the analyses described below.
Population structure and phylogenetic analyses
Population structure was characterised using: (1) individual distance-based phylogenetic analysis, (2) model-based clustering and (3) assumption-free Principal Component Analysis (PCA). To ensure that analyses would not be distorted by the presence of SNPs in a strong linkage disequilibrium (LD), the --indep command in PLINK was used to prune the SNPs that passed the initial filtering step. This was achieved by removing one locus from each pair for which LD (r 2 ) exceeded 0.1 within 50 SNP blocks resulting in 16,645 remaining SNPs. To estimate and test the phylogenetic relationship of different breeds we constructed a neighbour-joining (NJ) tree (Saitou and Nei 1987) based on individual genotypes in FastNJ software (Li 2015). Tree topology was tested with 1000 bootstrap replications. Nodes with <70% support were collapsed and the resulting tree was visualised using FigTree software (Rambaut and Drummond 2012). To evaluate the fractions of putative ancient populations in the modern genetic pool we used the fastSTRUCTURE (v1.0) clustering and stratification program (Raj et al. 2014). The program runs were carried out assuming between one and 40 groups (K) for both global set and Russian breed (including also closely related world breeds with more than five sampled individuals identified from the global NJ tree) sets. The cluster membership matrices of the fastSTRUCTURE outputs were visualised using PONG software (Behr et al. 2016). We used model complexity that maximises marginal likelihood to infer the putative optimal number of genetic clusters. As an assumption-free illustration of the differentiation between breeds, PCA, was performed using the SNPrelate Bioconductor package (Zheng et al. 2012).
Single nucleotide polymorphism diversity (SNP), linkage disequilibrium (LD), and haplotype sharing
An estimate of expected heterozygosity (H e ) at each locus was calculated using the --hardy command in PLINK and the mean value was calculated for each breed. The proportion of polymorphic loci (P n ) in each breed and the mean inbreeding coefficient (F) values were calculated using the PLINK commands --freq and --het, respectively. To calculate pairwise differentiation (F ST) between different breeds we used smartpca software from the Eigensoft package (v 6.1.4) (Patterson et al. 2006).
Runs of homozygosity (ROH) represent long stretches of haplotypes identical by descent (IBD) and provide valuable information about past and recent demographic events which accompanied the history of populations. To calculate ROH we used the methodology of Purfield et al. (2012) with stringent settings suitable for low-density genotype samples: ≥1 SNP per 80 Kbp region, >30 SNPs per region, with no more than one heterozygous SNP (PLINK commands: --homozyg-density 80 --homozyg-snp 30 --homozyg-het 1). To investigate the relationship and to infer signatures of recent gene flows between pairs of populations we used the method based on the detection of IBD-shared haplotypes according to Ralph and Coop (2013). Briefly, the genotypes of the global dataset were split by chromosome and phased using SHAPEIT 2 software (Delaneau et al. 2013) with 400 conditioning states (--states 400) and the effective population size (N e ) equal 15,000 as a safe provisional estimate for our diverse dataset. We used a high-density genetic map of the cattle genome (Ma et al. 2015) to correct for local variations in recombination rate during the haplotype inference. The haplotype sharing analysis was conducted using BEAGLE 4.1 software (Browning and Browning 2013) based on phased haplotypes with LOD score ≥ 2.5 (ibdlod = 2.5), the length of shared haplotypes ≥ 100 Kbp (ibdcm = 0.01) and the number of markers trimmed from the end of the shared haplotypes when testing for IBD equalled three (ibdtrim = 3). The inferred shared haplotypes were binned into three categories according to the size (<3 Mbp, 3–5 Mbp, >7 Mbp) and plotted using the R libraries igraph and ggcorrplot (R Development Core Team 2008).
To further reveal traces of genetic admixtures and their directions between the Russian and closely related breeds, and between the Russian and breeds of B. indicus origin, we applied the maximum-likelihood algorithm implemented in the Treemix software (Pickrell and Pritchard 2012) which models migration events on the phylogenetic tree. Two datasets analysed separately were: (1) Russian and closely related world breeds as defined from the NJ analysis, (2) Russian breeds and breeds of known B. indicus and B. javanicus origin from Decker et al. (2014) with at least five sampled individuals per breed. The Treemix analysis was performed with 1 SNP per block for estimation of the covariance matrix (k = 1) and gradual addition from one to 15 migration events with the step equal to one for the first dataset and from three to 18 migration events with the step equal to three for the second dataset. We rooted the trees on the Yakut cattle and B. javanicus for the first and second datasets, respectively. The optimal number of migration events was determined after examining the difference between the likelihoods of the tree after each migration step being added and the tree’s previous step likelihood (Δ Likelihood).
To estimate the historical and recent effective population sizes (N e ) in Russian breeds we applied a method based on the relationship between the extent of LD, N e , and the recombination rate within the populations implemented in the SNeP software (Barbato et al. 2015). The calculations were performed on SNPs with MAF≥0.05, with sample size correction (samplesize), and with minimum and maximum distances equal to 5000 bp and 2,000,000 bp, respectively. The recombination correction was applied according to Sved and Feldman (1973). LD values for size bins in the range from 28 to 600 Kbp were extracted from the SNeP output and plotted to estimate the LD decay for Russian cattle breeds.
Results
SNP, diversity, inbreeding, and LD within the Russian breeds
Both the GGP HD150K and BovineSNP50K SNP arrays were found highly informative for the Russian cattle breeds (Supplementary Table 1). The proportion of loci polymorphic (P n ) in at least one breed for the overlapping set of 26,701 SNPs shared between the arrays varied from 0.650 for the Red Pied to 0.977 for the Black Pied breeds with a mean of 0.891 (Table 1). The mean MAF was found highly consistent among the breeds ranging from 0.205 (Yakut) to 0.269 (Black Pied and Kalmyk). Similarly, the expected heterozygosity (H e ) was relatively high in all Russian breeds (range 0.271–0.352, mean 0.324) with the lowest values observed in the Red Pied (0.271) and Yakut (0.273) and the highest in the Black Pied (0.352) and Tagil (0.350). The inbreeding coefficient (F) demonstrated negative values for all the breeds, but the largest deviations from zero (>0.1) should be taken with caution because they were observed for the breeds with the lowest number of samples analysed (i.e., Istoben, Red Pied, Red Steppe, and Yurino; Table 1) suggesting that the genetic composition was likely not covered in full for these breeds.
We estimated the recent and past effective population (N e ) sizes for the native breeds and plotted the results (Supplementary Fig. 1). All of the Russian breeds demonstrated a highly similar pattern of N e decay with an increased rate starting ~200 generations ago (Supplementary Fig. 1b) likely being caused by bottlenecks associated with contemporary breed formation. The highest historical N e sizes were observed for Buryat and Kalmyk and the lowest N e for Yakut (Supplementary Fig. 1a,b). The LD decay plot (Fig. 1) suggested the presence of long haplotypes usually associated with low N e size (e.g., Yakut, Kostroma and Kholmogory); the most pronounced effect was observed for the Yakut cattle. The Buryat and Kalmyk cattle demonstrated a rapid LD decay consistent with the historically larger N e sizes of these breeds.
Consistent with the expectation of high inbreeding within Yakut, Kostroma, Kazakh Whiteheaded and Ukrainian Grey breeds, the presence of the longest and most frequent ROHs (>500 Kbp/>4 per animal) were observed within these breeds (Supplementary Figure 2). Tagil and Buryat demonstrated the shortest and the least frequent average number of ROHs in their genomes suggesting that these breeds could have been managed effectively to avoid excessive inbreeding. All other breeds expressed an intermediate level of ROHs consistent with the higher N e and expected moderate level of inbreeding.
Ancestry of Russian cattle breeds
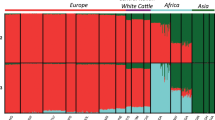
To identify ancestral relationships between native breeds from Russia and the cattle breeds distributed worldwide, we analysed our datasets with that of world breeds (Decker et al. 2014) and additional Eurasian breeds (Iso-Touru et al. 2016). As expected, the first two components of PCA differentiated the main clusters of breeds from Africa, Asia, and Europe representing mainly African taurine, cattle of Eurasian taurine origin and cattle of Asian indicine origin, with breeds expressing various levels of hybridisation found in between (Supplementary Fig. 3). Breeds from the Americas clustered with the European and Asian breeds. The majority of Russian breeds followed the European taurine breed cluster with additional breeds found in the cluster of taurine Asian breeds (Supplementary Fig. 3). These results were highly consistent with the fastSTRUCTURE analysis which suggested a close relationship between the breeds from Russia and other taurine breeds of European and several of Asian origin (Fig. 2). However, at K = 4 a separate cluster was formed by the Yakut cattle. The next breed that formed a separate cluster was the British Shorthorn (K = 5).
The collapsed NJ tree grouped samples into the well-supported breed-specific nodes confirming the expected phylogenetic relationships within the breed populations (Fig. 3). The three major well-resolved branches of the tree separated the breeds of Asian, African and European/American origins consistent with the PCA and fastSTRUCTURE results. The majority of the breeds from Russia (N = 17) were distributed along the branch of the European/American taurine breeds with some of them forming well-supported clusters with other breeds indicating close relationships. The Yakut cattle was found in the same cluster with Hanwoo and Wagyu cattle from Korea and Japan, respectively, near the Buryat cattle node. Other two well-resolved clusters involving Russian breeds and world breeds have grouped together the Kazakh Whiteheaded breed from Russia with Hereford samples from Russia and Wales; and the Ala-Tau and Kostroma with two breeds of European origin (Braunveih and Brown Swiss). Ukrainian Whiteheaded, Gorbatov Red and Istoben formed a separate cluster on the branch of the European breeds. Yurino formed a cluster with the Pinzgauer cattle from Austria. Kholmogory, Black Pied, Tagil and Red Steppe formed a large cluster with the Holstein-Friesian, French Red Pied and Lithuanian Light Grey breeds.
The nodes with less than 70% bootstrap support were collapsed. Yellow—Asian cattle (predominately B. indicus), green—African cattle (predominantly, taurines) blue—American cattle, brown—European cattle, red—Russian cattle. The names of Russian and their sister breeds from other regions are shown. In bold are names of the Russian cattle breeds shown on the images
The pairwise analysis of shared haplotypes between the Russian cattle breeds and taurine breeds of European and Asian origins has identified 39 breeds with a significant level of haplotype sharing with at least one breed from Russia (LOD > 2.5; Fig. 4, Supplementary Fig. 4). The top 10 world breeds that shared haplotypes at all three levels of the haplotype analysis were: Brown Swiss, Hereford, Holstein, Braunvieh, Senepol, French Red Pied, Beef Shorthorn, Maine Anjou, Norwegian Red, and Jersey (Fig. 4, Supplementary Fig. 4). This analysis has provided evidence for additional breed relations to the previously described results, and allowed us to distinguish between older and more recent relationships. Sharing of the short haplotypes (0–3 Mbp; presumably indicative of older relationships between populations) has formed two clear large clusters and two smaller clusters of breeds (Fig. 4a). The largest cluster revealed the ancestral relationships between the Northern French, British, and Finnish breeds with the Yaroslavl, Bestuzhev, Black Pied, Tagil, and Kazakh Whiteheaded breeds from our set. The second large cluster suggested further ancestral relationships between the South-European breeds from South-East France, Italy, Switzerland with Kostroma and Ala-Tau breeds from our dataset. Ukrainian Grey cattle samples both from Russia and Serbia shared short haplotypes with Podolian cattle (Serbia) and Romagnola (Italy) breeds. The Yakut and Buryat breeds formed a separate small cluster with Japanese Wagyu cattle whereas the Kalmyk cattle had significant haplotype sharing only with the Beef Shorthorn from England.
The longest shared haplotypes (>7 Mbp; likely indicative of recent introgression and admixture events, Fig. 4b) revealed the recent admixture between the Ukrainian Grey sampled in Russia (Boussaha et al. 2015) and the Yakut cattle. The Ukrainian Grey breed sampled in Serbia (Iso-Touru et al. 2016) did not demonstrate this pattern. Both the Kostroma and Ala-Tau breeds had extensive haplotype sharing with the Brown Swiss and Braunvieh. Multiple Russian breeds (i.e., Bestuzhev, Black Pied, Tagil, Yaroslavl, Kholmogory) shared haplotypes with Holstein-Friesian, Senepol, French Red Pied Lowland and Normande breeds.
To investigate more closely the genetic ancestry of the sampled Turano-Mongolian breeds (Yakut, Buryat, Kalmyk) we plotted the extent of pairwise haplotype sharing for each breed from highest to lowest value (Fig. 5 and Supplementary Fig. 5) for shortest haplotype segments (0–3 Mbp) including both taurine and indicine world breeds. The closest breeds (>1.5 Mbp average total haplotype length shared per animal) to Yakut cattle were Hanwoo, Buryat, Wagyu, Qinchan, Mongolian cattle and Morucha demonstrating a pronounced signal on the plot. The Buryat breed was mostly related to Wagyu, Hanwoo, Yakut, Qinchuan, Ala-Tau and Mongolian cattle breeds. Thus, Yakut and Buryat breeds showed a close relationship with taurine Asian breeds (and with each other) confirming their shared ancestry. Our samples of the Kalmyk cattle demonstrated mostly low values of haplotype sharing with the strongest relationship to Beef Shorthorn and a much weaker sharing with Wagyu and Welsh Black breeds. Interestingly, another sampling of the Kalmyk breed (Iso-Touru et al. 2016) showed some higher signal values, although it confirmed a relationship between the Kalmyk cattle and both taurine Asian (Hanwoo) and European (Simmental, Beef Shorthorn, Groningen Whitehead) breeds (Supplementary Fig. 5). In our analysis breeds of known indicine origin did not demonstrate high degree of haplotype sharing with Turano-Mongolian breeds with signal values always much lower than the values observed for the top taurine breeds (Fig. 5 and Supplementary Fig. 5).
Vertical lines indicate positons of B. indicus breeds. Sharing with the Ukrainian Grey was removed from the Yakut breed pairwise comparison due to pronounced signature of a very recent introgression from the Yakut breed, not identified for the Ukrainian Grey samples collected in Serbia. Breed names are shown for the largest number of shared haplotypes (>1.5 Mbp)
The relationships between the cattle breeds from Russia and the closely related world breeds
To reveal the fine-structure relationship between the Russian cattle breeds and the set of closely related world breeds we performed a separate PCA and a fine-scale admixture analysis of the breeds from Russia and eight world breeds that formed well supported clusters with the Russian breeds on the collapsed NJ tree (see Fig. 3). The first two components of PCA revealed four major clusters of the breeds (Supplementary Fig. 6a). The largest cluster contained the Holstein-Friesian cattle with the Black Pied and other European and Russian breeds that likely had been influenced by European dairy cattle genetics during their formation; the second cluster combined the Buryat, Kalmyk, Ukrainian Grey and Asian taurine breeds (Hanwoo and Wagyu). Another cluster combined the Kazakh Whiteheaded, Hereford cattle and the last cluster was formed by the Yakut cattle alone. The third principal component of PCA separated the Ukrainian Grey cattle from the cluster of the taurine breeds of Asian origin and revealed a separate cluster formed by the Kostroma, Brown Swiss, Ala-Tau and Braunvieh breeds (Supplementary Fig. 6b). The most likely number of populations according to the maximum likelihood estimation was equal to 11 (Supplementary Fig. 7). The fineSTRUCTURE results (Fig. 6) suggest that the most distant breeds within this set were the Yakut and Kholmogory, separated from the other breeds at K = 2, followed by the Hereford and Kazakh Whiteheaded group at (K = 3). The next cluster was formed by Kostroma and Brown Swiss (K = 4) followed by the Ukrainian Grey cattle (K = 5). At K = 6 it becomes apparent that the Asian taurine breeds (Hanwoo and Wagyu) cluster with the Kalmyk and Buryat cattle; the genetic material represented by the Hanwoo had a larger contribution on the Kalmyk and Buryat breeds than on Wagyu. At K = 7 a central cluster of mostly composite breeds with the influence of Holstein-Friesian/Black Pied genetic material becomes apparent with the Yaroslavl separating from this cluster at K = 10. We cannot exclude the possibility that the number of optimal genetic clusters in our analysis has been influenced by the unequal breed sample size and, in particular, by a small number of individuals collected for the Yurino, Red Steppe, Red Pied, Gorbatov Red and Istoben breeds. A larger number of samples would be needed to confirm the genetic composition of these breeds.
The Treemix results for the Russian cattle and most related other breeds (Supplementary Fig. 8) demonstrated the highest Δ likelihood increase for two migration events: the first one from the Yakut to the Ukrainian Grey breed collected in Russia and the second one from the Holstein to Tagil breed. Both results were in agreement with the observations made based on the haplotype sharing. For the Russian breeds combined with known B. indicus breed set (Supplementary Fig. 9), the highest gain in likelihood was received for nine migration events without any of them suggesting migration links between the Russian and indicine cattle populations.
The analysis of the F ST distances between the Russian breeds and those breeds closely related to them (Supplementary Table 2) revealed a low level of genetic differentiation with the mean value equal to 0.096 and a range from 0.003 to 0.235. The strongest differentiation involving a Russian breed was observed between the Podolian and Yakut cattle breeds while the lowest values were observed between the Red Pied and Finnish Ayrshire (F ST = 0.003). The Red Pied breed had a very low number of samples in our dataset (N = 2, Table 1) and these results should be taken with caution. Apart from this, the lowest F ST values were found between the samples of the Yaroslavl breed collected by us and by Iso-Touru et al. (2016). Surprisingly, the differentiation between the Black Pied and Holsten breeds (F ST = 0.020) was lower than the F ST observed between the Hereford samples from Russia and Wales (F ST = 0.029). The Yakut cattle has consistently demonstrated higher F ST values with other breeds, with the lowest differentiation observed with the Buryat cattle followed by the Kalmyk and Hanwoo breeds. Interestingly, the Wagyu breed, which had a high fraction of haplotypes shared with the Yakut cattle and was found next to it on the phylogenetic tree, had one of the highest degrees of differentiation with it (F ST = 0.20), following the Hereford and Podolian cattle.
Discussion
The advent of cost-efficient genotyping SNP arrays has made it possible to reveal the genetic profiles of various breeds of domesticated species, develop informed strategies of their improvement on one hand, and learn about the genetic processes accompanying domestication and breed formation on the other. While most efforts are dedicated to studying popular commercial breeds, e.g., Texel in sheep (Mucha et al. 2015) and Holstein-Friesian in cattle (van Binsbergen et al. 2015), there is a growing interest in the genetics of smaller local breeds because of the unique adaptations found in their genomes and their potential to contribute to solving problems in agriculture related to environmental change (e.g., global warming) and local pathogen resistance (Beynon et al. 2015). To this end we performed genotyping of 18 cattle breeds bred in Russia selected on the basis of a likely historical contribution of local cattle populations onto their contemporary genomes and compared them to commercial and native breeds previously collected from around the world (Decker et al. 2014; Iso-Touru et al. 2016). Along with the highly popular abundant Russian breeds (e.g., Black Pied or Kholmogory) we included highly specialised breeds that demonstrate extensive adaptations to specific environments (e.g., Yakut) and/or were almost extinct (e.g., Buryat). Therefore, our current dataset represents the largest and most complete set of the cattle breeds from Russia available for population genetic studies so far.
In agreement with the geographical position of Russia and its historical and trade links, the majority of the Russian cattle breeds demonstrated extensive common ancestry with the taurine cattle breeds from Europe. As expected to result from the ‘uncontrolled’ and/or complex breeding strategies started as early as in the 18th century (Dmitriev and Ernst 1989), for most of the Russian breeds we could not clearly identify their sister foreign breeds on the phylogenetic tree, except for the European and the Russian cattle being found on the same wide phylogenetic node. However, there were several examples where our data have confirmed the known historical relationships among the Russian cattle and some foreign breeds demonstrating robustness of our results. The most profound of these links is between the Kazakh Whiteheaded and Hereford breeds from both Russia and Europe, well supported by the known recent breeding history of the Kazakh Whiteheaded. The breed was formed between 1930 and 1950, by crossing of the Turano-Mongolian Kazakh and Kalmyk cattle with Hereford in the Kazakh Republic of the USSR (Dmitriev and Ernst 1989). Another example of known relations and historical breed formation (Dmitriev and Ernst 1989) was confirmed by clustering of Kostroma, Brown Swiss, Braunveih and Ala-Tau consistently supported by the structure, phylogenetic, haplotype analysis and population differentiation levels (F ST ranges 0.032–0.069). A separate node on our phylogenetic tree formed by the Kholmogory, Holstein-Friesian, Black Pied and several related European breeds further supported by haplotype sharing likely reflects the historical relations that trace back to the 17th century when the Kholmogory breed was formed and later interbred with “Dutch cattle” (Dmitriev and Ernst 1989). However, the structure analysis indicates that the genetic component of the contemporary Holstein-Friesian breed in Kholmogory is relatively small and that Kholmogory should be considered genetically distinct, supporting previous observations (Zinovieva et al. 2016). On the other hand, our samples of the Russian Black Pied breed demonstrate a very low differentiation from the Holstein-Friesian (F ST = 0.02, e.g., lower than between the two sets of Hereford samples in our analysis) suggesting that the use of imported Holstein-Friesian sires/semen in Russia could have significantly affected the Black Pied’s genetics. Haplotype sharing analysis that was based on short haplotype blocks (presumably reflecting ancestral relationships) has further confirmed a complex history of the Russian cattle breeds of European origin but has allowed us to assign them to three major clusters based on predominantly shared haplotypes. While the largest cluster mostly contained Russian breeds with historical influence from highly commercial European breeds (e.g., Holstein-Friesian and Angus) and other breeds that could also have been influenced by these multinational breeds, the second one was built around the related Kostroma, Brown Swiss, Braunveih and Ala-Tau breeds with the addition of several other breeds from France, Italy and Germany. The Ukrainian Grey cattle formed the last separate cluster shared only with the Podolian and Romadnola breeds confirming the Ukrainian Grey’s position within the primitive Podolian group of cattle breeds (Kushnir and Glazko 2009).
In addition to extensive links to cattle of European ancestry, PCA suggested that there are breeds in Russia that have shared ancestry with cattle from Asia. In agreement with this, the Yakut, Buryat, and Kalmyk cattle clustered with the Turano-Mongolian and other Asian taurine breeds on the phylogenetic tree and structure plots. While on the structure global dataset the Yakut breed formed the first breed-specific cluster after the observed divergence of B. indicus and African taurines, on the phylogenetic tree, it was found on the same node with Buryat cattle and other taurine Asian breeds. The exact reason for the Yakut cattle being so divergent based on structure results is not currently clear but could be related to a combination of its low historical N e combined with long isolation from other breeds. A closer relation of the Yakut cattle with other divergent Asian Turano-Mongolian breeds may imply their early separation from the rest of the taurine gene pool or even independent domestication in Asia (Mannen et al. 2004). Haplotype sharing results further confirm these relationships within the Turano-Mongolian breed set placing the Yakut cattle on the same cluster with Buryat and Wagyu and indicating links with Hanwoo and the Mongolian cattle. The Buryat cattle was considered to be extinct until quite recently when a herd was discovered in Mongolia and imported back to Russia to start recovering the breed. Our results indeed demonstrated that the Buryat shares more haplotypes with Yakut and Wagyu cattle than with the breeds from Mongolia suggesting its separate origin from Mongolian cattle. Interestingly, the placement of the third Turano-Mongolian breed on our list, the Kalmyk, remains unclear. While it formed a separate cluster within the European cattle on the phylogenetic tree, structure results suggested a common ancestry with Buryat and Hanwoo breeds. Haplotype sharing showed a strong recent admixture with the Beef Shorthorn. The latter can be explained by the known use of Shorthorn to ‘improve’ the Kalmyk cattle in the USSR (Dmitriev and Ernst 1989). This likely had an impact on the genetics of this breed and affected its position on the phylogenetic tree masking the expected ancestral relationships that were picked up only by the structure analysis. Another example of a likely effect of a recent admixture on the genetics of a breed was observed during comparison of the Ukrainian Grey cattle samples originating from Serbia (Iso-Touru et al. 2016) and from Russia (Boussaha et al. 2015). While these sample sets cluster together suggesting that they indeed belong to the same breed, the sample set from Russia demonstrated a clear evidence of a recent admixture with the Yakut cattle based on the haplotype sharing and confirmed by Treemix analysis. The samples from Serbia had no traces of this event.
Interestingly, we did not identify any significant evidence of admixture between any of the Russian breeds and the indicine cattle neither in haplotype sharing nor the Treemix analyses. However, the structure global plot (K = 3) suggested some level of indicine ancestry in the Turano-Mongolian breeds. This observation may imply a very ancient and probably weak admixture event not detected by other methods. It is also possible that the BovineSNP50K array SNP loci (and, as a result, the set of SNPs used in the present work) bias to taurine and ancient SNPs shared by taurine and indicine populations (McKay et al. 2008) has affected our results to some extent and masked admixture with B. indicus. Both scenarios suggest that more detailed studies involving the whole-genome resequencing of Russian cattle genomes and their comparison to both the taurine and indicine genome references would be needed to resolve this issue and shed additional light on the reasons for observed divergence of the Yakut cattle.
When analysed individually or in the context of only the most related world breeds, the Russian cattle breeds demonstrated a modest level of genetic diversity and comparable estimates of effective population sizes with other Eurasian breeds (Iso-Touru et al. 2016). The Kholmogory and Yakut breeds are further confirmed as the most genetically distinct within the set of the breeds from Russia and related Eurasian breeds on the structure results, supported by high F ST . Strong influence of Holstein-Friesian genetics became apparent in a separate cluster of breeds. Yaroslavl breed separated from this cluster at K = 10 being the last Russian cattle breed that demonstrated unique genetics while other breeds including the Black Pied, Tagil, Bestuzhev, Istoben, Yurino, and Ukrainian Whiteheaded demonstrated different levels of Holstein-Friesian contribution to their genetics suggesting that these breeds might have left with a relatively small fraction of alleles from native populations. This was supported by a relatively low level of population differentiation within this group (F ST range 0.020–0.094). However, we cannot exclude that both the SNP loci bias to a small number of taurine breeds and small sampling sizes for some breeds in our list could have influenced these results.
The presence of long runs of homozygosity in the Yakut, Kostroma and Ukrainian Grey breeds might indicate either a high level of adaptation and specialisation or effects of inbreeding and low effective population size. Regardless of the reason, this information should be considered during the development of breeding programs for these populations. The genetic uniqueness of the highly adapted to harsh climatic conditions Yakut breed should stimulate and guide its recovery program.
Herein we provide the first detailed view on the population genetics of a comprehensive list of the cattle breeds bred in Russia that potentially have arisen from local cattle populations and/or could be adapted to harsh environments and climate. Our results demonstrate that some of the breeds studied have distinct genetic profiles (e.g., Kholmogory, Yakut, Yaroslavl) making them priority targets for deeper studies to reveal signatures of selection and adaptations related to local environments and for conservation purposes. We also observed that a large group of breeds had both old and recent influence from commercial European breeds (e.g., Kostroma, Kazakh Whiteheaded, Istoben) meaning that their genomes could potentially contain only a small fraction of ancestral alleles, but these could be important for surviving local conditions and can be used for admixture mapping programs aiming at economically important traits (Kassahun et al. 2015). The links between the Russian breeds and breeds from other countries presented in this study form a basis for future work on contrasting their genomes to reveal causative alleles or haplotypes using a right set of related and outgroup populations for the comparison to avoid ‘signal dilution’ or false positive signals. The uniqueness of the Yakut breed shown in this study makes it a priority for further detailed studies on one hand, but makes it difficult to identify the right breeds to contrast it to on the other; implying that additional, more detailed studies of Russian native cattle breeds of Asian origin in the context of a larger set of Asian taurine and indicine breeds might be required to fully benefit from their unique genetics.
Data Archiving
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.68hv7
References
Barbato M, Orozco-terWengel P, Tapio M, Bruford MW (2015) SNeP: a tool to estimate trends in recent effective population size trajectories using genome-wide SNPdata. Front Genet 6:109
Behr AA, Liu KZ, Liu-Fang G, Nakka P, Ramachandran S (2016) pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 32(18):2817–2823
Beynon SE, Slavov GT, Farre M, Sunduimijid B, Waddams K, Davies B et al (2015) Population structure and history of the Welsh sheep breeds determined by whole genome genotyping. BMC Genet 16:65
Boichard D, Brochard M (2012) New phenotypes for new breeding goals in dairy cattle. Animal 6(4):544–550
Boussaha M, Letaief R, Yudin N, Voevoda M, Rocha D (2015) The extent of linkage disequilibrium in the ancient Ukrainian grey cattle breed. Glob J Anim Breed Genet 3(4):159–163
Bovine HapMap Consortium (2009) Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324(5926):528–532
Browning BL, Browning SR (2013) Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194(2):459–471
Buchanan DS, Lenstra JA (2015) Breeds of Cattle. Genetics of Cattle. 2nd edn.: 33-66
DAD-IS (2017) Vol. 2017. Food and Agriculture Organization of the United Nations, Rome, Italy
Decker JE, McKay SD, Rolf MM, Kim J, Molina Alcala A, Sonstegard TS et al. (2014) Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet 10(3):e1004254
Delaneau O, Zagury JF, Marchini J (2013) Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 10(1):5–6
Dmitriev NG, Ernst LK (1989) Animal genetics resources of the USSR. Food and Agriculture Organization of the United Nations, Rome, Italy
Dunin IM, Dankvert AG (2013) Breeds and types of farm animals in the Russian Federation. Ministry of Agriculture, Moscow, Russian Federation
Edwards CJ, Ginja C, Kantanen J, Perez-Pardal L, Tresset A, Stock F et al. (2011) Dual origins of dairy cattle farming--evidence from a comprehensive survey of European Y-chromosomal variation. PLoS ONE 6(1):e15922
Gao Y, Wu H, Wang Y, Liu X, Chen L, Li Q et al. (2017) Single Cas9 nickase induced generation of NRAMP1 knockin cattle with reduced off-target effects. Genome Biol 18(1):13
Gaouar SB, Da Silva A, Ciani E, Kdidi S, Aouissat M, Dhimi L et al. (2015) Admixture and local breed marginalization threaten Algerian sheep diversity. PLoS ONE 10(4):e0122667
Iso-Touru T, Tapio M, Vilkki J, Kiseleva T, Ammosov I, Ivanova Z et al. (2016) Genetic diversity and genomic signatures of selection among cattle breeds from Siberia, eastern and northern Europe. Anim Genet 47(6):647–657
Kassahun Y, Mattiangeli V, Ameni G, Hailu E, Aseffa A, Young DB et al. (2015) Admixture mapping of tuberculosis and pigmentation-related traits in an African-European hybrid cattle population. Front Genet 6:210
Kushnir AV, Glazko VI (2009) Gray Ukrainian cattle and their closely related forms. Contemp Probl Ecol 2(3):288–295
Larkin DM, Yudin N (2016) The genomes and history of domestic animals. Mol Genet Microbiol Virol 31(4):197–202
Li JF (2015) A fast neighbor joining method. Genet Mol Res 14(3):8733–8743
Li MH, Kantanen J (2010) Genetic structure of Eurasian cattle (Bos taurus) based on microsatellites: clarification for their breed classification. Anim Genet 41(2):150–158
Loftus RT, MacHugh DE, Bradley DG, Sharp PM, Cunningham P (1994) Evidence for two independent domestications of cattle. Proc Natl Acad Sci U S A 91(7):2757–2761
Ma L, O’Connell JR, VanRaden PM, Shen B, Padhi A, Sun C et al. (2015) Cattle sex-specific recombination and genetic control from a large pedigree analysis. PLoS Genet 11(11):e1005387
Mannen H, Kohno M, Nagata Y, Tsuji S, Bradley DG, Yeo JS et al. (2004) Independent mitochondrial origin and historical genetic differentiation in North Eastern Asian cattle. Mol Phylogenet Evol 32(2):539–544
Mason IL (1969) A World dictionary of livestock breed types and varieties commonwealth agricultural bureaux. Farnham Royal, UK
Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP et al. (2009) Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 4(4):e5350
McKay SD, Schnabel RD, Murdoch BM, Matukumalli LK, Aerts J, Coppieters W et al. (2008) An assessment of population structure in eight breeds of cattle using a whole genome SNP panel. BMC Genet 20(9):37
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16(3):1215
Mucha S, Bunger L, Conington J (2015) Genome-wide association study of footrot in Texel sheep. Genet Sel Evol 47:35
Nicolazzi EL, Caprera A, Nazzicari N, Cozzi P, Strozzi F, Lawley C et al. (2015) SNPchiMp v.3: integrating and standardizing single nucleotide polymorphism data for livestock species. BMC Genom 16:283
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2(12):e190
Pickrell JK, Pritchard JK (2012) Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8(11):e1002967
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575
Purfield DC, Berry DP, McParland S, Bradley DG (2012) Runs of homozygosity and population history in cattle. BMC Genet 13:70
Raj A, Stephens M, Pritchard JK (2014) fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197(2):573–589
Ralph P, Coop G (2013) The geography of recent genetic ancestry across Europe. PLoS Biol 11(5):e1001555
Rambaut A, Drummond AJ (2012). FigTree. Version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree
R Development Core Team (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sambrook J, Russell DW (2006) The condensed protocols from Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA
Sved JA, Feldman MW (1973) Correlation and probability methods for one and two loci. Theor Popul Biol 4(1):129–132
Upadhyay MR, Chen W, Lenstra JA, Goderie CR, MacHugh DE, Park SD et al. (2017) Genetic origin, admixture and population history of aurochs (Bos primigenius) and primitive European cattle. Heredity 118(2):169–176
van Binsbergen R, Calus MP, Bink MC, van Eeuwijk FA, Schrooten C, Veerkamp RF (2015) Genomic prediction using imputed whole-genome sequence data in Holstein-Friesian cattle. Genet Sel Evol 47:71
Yudin NS, Vasil’eva LA, Belyavskaya VA, Aitnazarov RB, Smirnov PN, Heaton M et al. (2015) Design of a DNA panel for genomic studies in Russian cattle breeds. Russian J Genet Appl Res 5(1):21–25
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28(24):3326–3328
Zinovieva NA, Dotsev AV, Sermyagin AA, Wimmers K, Reyer H, Sölkner J et al. (2016) Study of genetic diversity and population structure of five Russian cattle breeds using whole-genome SNPanalysis. Sel’skokhozyaistvennaya Biol 51(6):788–800
Acknowledgements
The work was supported by the Russian Science Foundation grant (RSF, 16–14–0090).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yurchenko, A., Yudin, N., Aitnazarov, R. et al. Genome-wide genotyping uncovers genetic profiles and history of the Russian cattle breeds. Heredity 120, 125–137 (2018). https://doi.org/10.1038/s41437-017-0024-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-017-0024-3
This article is cited by
-
On the origin and diversification of Podolian cattle breeds: testing scenarios of European colonization using genome-wide SNP data
Genetics Selection Evolution (2021)
-
The genetic heritage of Alpine local cattle breeds using genomic SNP data
Genetics Selection Evolution (2020)
-
Assessing the power of principal components and wright’s fixation index analyzes applied to reveal the genome-wide genetic differences between herds of Holstein cows
BMC Genetics (2020)
-
Refining the genetic structure and relationships of European cattle breeds through meta-analysis of worldwide genomic SNP data, focusing on Italian cattle
Scientific Reports (2020)
-
Intrabreed and interbreed variation of the BOLA-DRB3.2 gene in the Kostroma and Yaroslavl indigenous Russian cattle breeds
Immunogenetics (2020)