Abstract
Purpose
Evaluate whether fragile X syndrome (FXS) testing should be transitioned to a second-tier test in global developmental delay, intellectual disability, and autism spectrum disorder in the absence of family history and suggestive clinical features.
Methods
Determine the diagnostic yield of FXS testing performed by the Alberta Children's Hospital (ACH) Molecular Diagnostic Laboratory between 2012 and 2017. Retrospective chart review of FXS-positive patients to determine presence or absence of suggestive clinical features and family history.
Results
Of the 2486 pediatric patients with neurodevelopmental disorders tested for FXS, 25 males and 5 females were positive. This corresponds to a 1.2% diagnostic yield of FXS testing at our center. Retrospective chart review of the FXS-positive cases revealed that 96% of FXS patients had either, if not both, clinical features or family history suggestive of FXS present at the time of testing. Only one patient had neither family history nor clinical features suggestive of FXS.
Conclusion
In 96% of FXS-positive cases, there was sufficient clinical suspicion raised on the basis of clinical features and/or family history to perform targeted FXS testing. We thus propose that in the absence of suggestive clinical features or family history, FXS testing should be transitioned to a second-tier test in neurodevelopmental disorders.
Similar content being viewed by others
INTRODUCTION
Fragile X syndrome (FXS) is an X-linked trinucleotide repeat disorder that has historically been referred to as the most common inherited cause of intellectual disability (ID).1,2 While the reported prevalence of this disorder varies throughout the literature, a meta-analysis from 2014 documented a frequency of 1.4 per 10,000 males and 0.9 per 10,000 females.3 This condition is caused by a loss-of-function variant of FMR1, which results in reduced or absent levels of fragile X mental retardation protein (FMRP).4 The decreased production of FMRP adversely impacts both pre- and postnatal neurodevelopment leading to a phenotype with a range of cognitive features including developmental delay, intellectual disability, and learning disability.
While FXS has historically been considered the most common inherited cause of ID, advances in genetic testing have elucidated a much wider variety of etiologies.1,5,6 The historical overrepresentation of FXS in the literature is largely a consequence of the previously limited molecular assays for the investigation of genetic causes of neurodevelopmental disorders. Advancements in next-generation sequencing (NGS) technology have revolutionized our clinical approach to genetic testing and have expanded our understanding of genetic causes of neurodevelopmental disorders. Fragile X testing is a single-gene molecular assay with a low diagnostic yield in this population around 1.5–2%.1 In comparison to this single-gene assay, the diagnostic yield of broader chromosomal microarray (CMA) is 15–20% and exome sequencing has a diagnostic yield of approximately 30%.1,7 Nonetheless, current recommendations from the American College of Medical Genetics and Genomics, the American Academy of Pediatrics, and the Canadian Paediatric Society suggest both FXS and CMA testing as first-tier investigations in neurodevelopmental disorders including global developmental delay (GDD), intellectual disability (ID), and autism spectrum disorder (ASD).5,8,9
There is an evolving culture shift within the fields of clinical genetics and developmental pediatrics that has produced growing evidence suggesting FXS testing may be more appropriate as a second-tier investigation in neurodevelopmental disorders. These studies have highlighted the low diagnostic yield of FXS testing in comparison with other testing modalities such as CMA and exome sequencing.1,6,10 Furthermore, evolving data suggest that rather than offering first-line FXS testing to all patients with a neurodevelopmental disorder, the FXS phenotype provides the opportunity to offer FXS molecular testing in targeted cases where clinical features or family history raise clinical suspicion for a diagnosis of FXS.1,6
The aim of our study is to evaluate whether fragile X testing should be transitioned to a second-tier test in global developmental delay, intellectual disability, and autism spectrum disorder in the absence of family history and suggestive clinical features. To address this clinical question, we studied our local population served by the Alberta Children’s Hospital’s Molecular Diagnostic Laboratory. We first hypothesize that our local review of the diagnostic yield of FXS testing in pediatric patients with neurodevelopmental disorders will be much lower than the established diagnostic yield of CMA and exome sequencing. Secondly, we hypothesize that individuals with positive FXS testing will have family history or clinical features suggestive of FXS at the time of testing.
MATERIALS AND METHODS
To address our hypotheses, we first calculated the diagnostic yield of FXS testing performed by our center. Second, we performed a retrospective chart review to determine the percentages of full mutation individuals with (1) clinical features suggestive of FXS, (2) family history suggestive of FXS, (3) clinical features and family history suggestive of FXS, and (4) neither clinical features nor family history suggestive of FXS. This study was approved by the Conjoint Health Research Ethics Board at the University of Calgary.
Population
We reviewed de-identified laboratory data from all pediatric patients with neurodevelopmental disorders tested for FXS by the Alberta Children’s Hospital’s Molecular Diagnostic Laboratory between 2012 and 2017. To meet the inclusion criteria, pediatric patients (0–18 years of age) required a diagnosis of ASD, ID, or GDD of unknown etiology that prompted FXS testing. Our population was comprised of 2486 patients, including 1919 males and 567 females.
Determining FXS testing yield
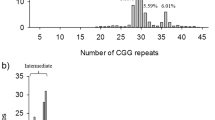
FXS testing results were classified as full mutation female, full mutation male, premutation female, premutation male, normal female, or normal male. Full mutation was considered a positive test result and refers to greater than 200 trinucleotide repeats, whereas premutation refers to 56–200 repeats, and normal refers to fewer than 56 repeats.4
Retrospective chart review
Subsequently, we performed a retrospective chart review of the 30 individuals who tested positive for FXS (full mutation and full mutation mosaic) during the six-year time period. We reviewed clinician notes and documentation from the appointment that prompted FXS testing to determine the presence or absence of clinical features suggestive of FXS. These clinical features include macrocephaly, elongated face, large ears, prognathism, and testicular enlargement.4 Documentation of any number of these clinical features or of an overall picture suggestive of FXS was considered to meet this criterion. Additionally, we reviewed the charts for family history of suspected X-linked inheritance, or for family history of confirmed or suspected FMR1-related disorders including fragile X syndrome, fragile X–associated tremor/ataxia syndrome, and FMR1-related primary ovarian insufficiency.
RESULTS
Our study included 2486 pediatric patients with neurodevelopment disorders who underwent FXS testing between 2012 and 2017 through the Alberta Children’s Hospital’s Molecular Diagnostic Laboratory for the purpose of identifying the underlying etiology of a neurodevelopmental disorder. Of these 2486 patients, only 25 males and 5 females were found to be positive for FXS with greater than 200 trinucleotide repeats on molecular testing. This corresponds to a diagnostic yield of 1.2% for FXS testing in our population.
Retrospective chart review of the 30 full mutation individuals who tested positive for FXS revealed that 26 patients had either, if not both, clinical features or family history suggestive of a FXS diagnosis prior to testing. In contrast, only one individual (a full mutation male) did not have clinical features or family history suggestive of a diagnosis of FXS. Data could not be obtained for three individuals as their charts were not available for review. Thus, of the 27 charts reviewed, a diagnosis of FXS could have been clinically suspected in 96% of patients on the basis of clinical features or family history.
The documentation of clinical features varied largely between clinicians, with some clinicians vaguely documenting “facial features consistent with FXS” and others reporting individual features, most commonly macrocephaly. If available in clinical documentation, a cutoff of head circumference greater than 2 standard deviations above the mean was used to identify macrocephaly. Notably, of the 19 total FXS patients with positive family history, 11 had a family history of FXS, 7 had a family history of primary ovarian insufficiency, and 1 had a family history of suspected X-linked ID.
DISCUSSION
In light of the historical overrepresentation of FXS as the most common inherited cause of ID prior to advances in NGS, FXS molecular testing is currently recommended as a first-tier investigation in neurodevelopmental disorders. Our study aimed to investigate the suitability of this first-tier test status. The results of this study support a transition towards second-tier testing in cases with low clinical suspicion for FXS as defined by absence of suggestive clinical features or family history. The yield of FXS testing performed by our molecular laboratory was 1.2% (Table 1), which is comparable with that reported in the literature of 1.5–2%. As hypothesized, the 1.2% yield of FXS testing performed by our center was much lower than the established yield of CMA and exome sequencing in neurodevelopmental disorders, 15–20% and 30%, respectively.1,7
Furthermore, we found that 96% (26/27) of patients who tested positive for FXS had at least one of, if not both, clinical features or family history suggestive of FXS (Table 2). Accordingly, in all but one of the positive FXS cases we reviewed, there was sufficient clinical suspicion on the basis of clinical features and/or family history to lead the clinician to perform FXS testing. In addition to the 27 FXS charts we reviewed, three charts were not available. We recognize that depending on these three patients, the percentage of FXS patients with either, if not both, clinical features or family history suggestive of FXS could have ranged from 86% (26/30) to 97% (29/30). Additionally, as we performed a retrospective chart review, our study is limited in the fact that there was no prospective planning of dysmorphology examination or documentation. However, this realistically represents variability in clinical practice. The findings ultimately make a strong case for targeted FXS testing founded on clinical suspicion raised by clinical features or family history, rather than FXS as a first-tier investigation for all individuals with neurodevelopmental disorders.
We acknowledge that targeted testing could risk missing patients with a more subtle FXS phenotype, especially in clinical centers less familiar with the FXS phenotype and other FMR1-related disorders. As such, we suggest transitioning FXS testing to a second-tier test in the absence of clinical features or family history suggestive of the diagnosis. With this approach, should a full mutation patient with a subtle phenotype not be tested for FXS in the first round of investigations, the appropriate diagnosis would be made through single-gene FXS testing included in the second-tier investigations. Although the diagnosis would not be missed, second-tier testing would result in a delay to diagnosis for the small percentage of FXS patients not initially tested based on clinical suspicion. The length of this delay to diagnosis would depend on a center’s turnaround time for first-tier investigations including NGS assays that do not detect triplet repeat disorder such as FXS, and on the time required for a patient to be seen in follow up. These delays are particularly important to consider in the context of establishing a diagnosis to prevent future affected pregnancies. Ultimately however, the transition of FXS testing to a second-tier test in the absence of suggestive clinical features or family history would avoid unnecessary testing without otherwise altering patient care.
Our study included 2486 individuals, which is among the largest populations examined to date in the context of similar studies performed by the Children’s National Health System Genetics Department in Washington (202 males tested with CMA and/or FXS testing), the Clinical Genetics Department at the Children’s Hospital of Eastern Ontario (1177 pediatric males tested for FXS), and the University of California–Los Angeles (UCLA) Medical Center (654 males tested for FXS).1,6,10 These studies report similarly low diagnostic yields of FXS testing in their populations, including 2.5% reported by the Weinstein et al. group in Washington, as high as 2.4% reported by the Hartley group from Ottawa, and 0% reported by the Mullegama et al. group from UCLA.1,6,10 By highlighting the low 1.2% local diagnostic yield of FXS testing in the context of our large study population, our study also draws attention to the issue of resource misallocation propagated by the current first-tier test status of single-gene FXS testing. Given the relatively high cost of genetic testing and the limited financial and personnel resources of any given diagnostic laboratory, transitioning FXS testing to second-tier would promote more efficient resource allocation within the health-care system.
As the field of clinical genetics continues to advance, further studies will need to be done to assess the yield of other assays including CMA and exome sequencing, and to determine which investigations should be considered first-line in neurodevelopmental disorders. Based on our findings, we advocate for a change in current practice guidelines and support the transition of single-gene FXS testing to a second-tier investigation in the identification of genetic etiology of neurodevelopmental disorders without clinical features or family history suggestive of FXS.
References
Weinstein V, Tanpaiboon P, Chapman KA, Mew NA, Hofherr S. Do the data really support ordering fragile X testing as a first-tier test without clinical features? Genet Med. 2017;19:1317–1322.
Gallagher A, Hallahan B. Fragile X-associated disorders: a clinical overview. J Neurol. 2012;259:401–413.
Hunter J, Rivero-Arias O, Angelov A, Kim E, Fotheringham I, Leal J. Epidemiology of fragile X syndrome: a systematic review and meta-analysis. Am J Med Genet. 2014;164A:1648–1658.
Monaghan KG, Lyon E, Spector EB. ACMG Standards and Guidelines for fragile X testing: a revision to the disease-specific supplements to the Standards and Guidelines for Clinical Genetics Laboratories of the American College of Medical Genetics and Genomics. Genet Med. 2013;15:575–586.
Belanger SA, Caron J. Evaluation of the child with global developmental delay and intellectual disability. Paediatr Child Health. 2018;23:403–410.
Hartley T, Potter R, Badalato L, Smith AC, Jarinova O, Boycott KM. Fragile X testing as a second-tier test. Genet Med. 2017;19:1380.
Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879.
Schaefer GB, Mendelsohn NJ. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15:399–407.
Moeschler JB, Shevell M. Comprehensive evaluation of the dhild with intellectual disability or dlobal developmental delays. Pediatrics. 2014;134:903–918.
Mullegama SV, Klein SD, Nguyen DC. Is it time to retire fragile X testing as a first-tier test for developmental delay, intellectual disability, and autism spectrum disorder? Genet Med. 2017;19:1380.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Borch, L.A., Parboosingh, J., Thomas, M.A. et al. Re-evaluating the first-tier status of fragile X testing in neurodevelopmental disorders. Genet Med 22, 1036–1039 (2020). https://doi.org/10.1038/s41436-020-0773-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0773-x