Abstract
Purpose
Summarize and interpret results from exercises distributed to laboratories offering cell-free (cf) DNA screening for Down syndrome.
Methods
The College of American Pathologists distributed three patient-derived plasma specimens twice in 2018. Sequencing platforms, test methods, results, and responses to supplemental questions were collected. Results were not graded but discrepancies were identified.
Results
Sixty-five laboratories from six continents enrolled; six provided no results. The most common methodology was shotgun/genome sequencing (39/56, 70%). Overall, 40% of the gestational or maternal age responses were incorrect but 45% of the errors were corrected by the next distribution. Fetal fractions from 54 responding laboratories generally agreed with the intended response. No genotyping errors occurred (40/40 for trisomy 21 and 226/226 for euploid challenges) but 10 additional tests failed (3.6%). All 213 fetal sex calls were correct. Participants reported their clinical text for a Down syndrome screen positive test; 39% were classified as inadequate or misleading.
Conclusion
Patient-derived materials are suitable for all enrolled technologies/methodologies, but collecting material is challenging. Suggested clinical text includes the terms “screen positive” and “screen negative.” Overall, laboratories performed well. Future efforts will focus on potential manufactured samples, clarifying results reporting and including additional chromosome abnormalities.
Similar content being viewed by others
INTRODUCTION
The discovery of cell-free (cf) DNA derived from the fetoplacental unit in maternal circulation was first reported in 19971 and the finding was initially used to identify fetal sex early in pregnancy.2 With the advent of next-generation sequencing (NGS), two groups3,4 reported proof of concept studies for identifying common autosomal trisomies in 2008. This led to the introduction of a clinical test for trisomy 21 in 2011.5 Since then, several testing methodologies have been demonstrated to be effective6 and large numbers of pregnancies have been screened globally. Such testing is often referred to as noninvasive prenatal testing (NIPT) or screening (NIPS), even though these terms also describe longstanding prenatal testing modalities such as serum screening. As NIPS usage expands, professional organizations and others have noted the lack of formal external proficiency testing (PT) schemes to assist in monitoring laboratory quality.7,8,9,10,11 One informal challenge demonstrated this frustration.12 Blood samples from postmenopausal, nonpregnant women were sent to five commercial NIPS laboratories, three of which reported results consistent with a female fetus. Despite flaws to that approach, the report drew attention to the need for more formal standards for process and review.13
Recently, two European groups (the United Kingdom National External Quality Assessment Scheme [UK NEQAS]14 and the European Molecular Genetics Quality Network [EMQN])15 published their findings from pilot trials.16 The first was based on manufactured samples that were expected to be suitable for most methodologies, except those employing single-nucleotide polymorphisms (SNPs). Of the 40 respondents, 10% reported genotyping errors, with an overall test failure rate of 18% and the material was judged unsuitable. The second pilot used patient-derived plasma and among 86 respondents, 2.3% made critical genotyping errors with an overall failure rate of 2.3%.
The College of American Pathologists (CAP) NIPT Educational Exercise was aimed at providing actual maternal plasma samples in 4-mL nonpooled aliquots to groups of laboratories. Use of lots allows for comparison of results among a group of participating laboratories (i.e., participants) receiving the same set of challenges. When referring to testing in general, the term “NIPS” is used, but when referring to the survey, “NIPT” is used. The aim was to document platforms and methodologies, fetal fraction methods and results, intermediate results (e.g., z-scores), clinical calls, and to request additional information concerning laboratory protocols through supplemental questions. A second aim was to validate the suitability of the plasma samples over a wide variety of NIPS methodologies. Although not a formal external proficiency testing program at this time, these exercises are part of the comprehensive CAP laboratory improvement activities that also include structured in-person laboratory inspections and formal laboratory accreditation.
MATERIALS AND METHODS
Preliminary activities
In 2012, the CAP modified its Molecular Pathology Checklist to include questions specific to NIPS. These assisted CAP inspectors in evaluating laboratories during the accreditation inspection process. In parallel, the development of an educational exercise for cfDNA testing of maternal blood to screen for Down syndrome was assigned to the CAP Biochemical and Molecular Genetics (BCMG) Committee. In 2013, pilot studies were initiated; the first using artificial samples and the second using pooled maternal plasma samples. The results confirmed prior assumptions that neither are suitable for all NIPS methodologies. A third pilot study using in-house control material from a laboratory that relied on the SNP methodology determined that this material was also not suitable. Based on these results, the BCMG concluded that at this time, actual nonpooled patient samples must be used to meet the requirements of all methodologies. This type of material was distributed for a final pilot study, which was successful. Based on these results, the Committee recommended moving forward with an educational exercise beginning in 2018. The results of the first year of that exercise form the basis of this report.
The NIPT educational exercise
The CAP offered the NIPT Educational Exercise for the first time in 2018.17 As NIPS is not regulated under CLIA regulations, the exercises were designed to provide two distributions (mailings) of three aliquots (4 mL each) of maternal plasma per year (challenges identified as NIPT-01 through 03 in the A distribution and NIPT-04 to 06 in the B distribution). The exercise is not a formal external proficiency testing program but an opportunity for laboratories around the world to compare methods, techniques, sample testing results, clinical interpretations, laboratory protocols, and epidemiological monitoring. As an educational activity, results were not graded, but discrepancies were identified so participants could take remedial action. Interpretations of the results of each exercise were reported promptly to registered participants.
Sample challenges
Blood samples were collected from donor pregnant women who were undergoing, or had undergone, NIPS for Down syndrome as part of routine prenatal care. Each was consented and asked to provide ten 10-mL aliquots of blood in Streck BCT® (La Vista, NE) containers. The tubes and associated information, including the clinical NIPS results, were sent to the CAP contracted processing center at Women & Infants Hospital of Rhode Island (WIH). No protected health information (PHI) was collected and the data were linked to the enrollment site only by a confidential subject ID. The enrollment sites received exemptions from their institutional research board (IRB) or equivalent indicating that these activities were not considered human research. The processing center at WIH also received IRB approval (1238505–1). Demographic information included enrollment site code, enrollment date, aneuploidy risk status (low, high), draw time, number of tubes, phlebotomist, date received, test results, time received, date processed, and temperature at receipt (cool or room). Test results included the NIPS clinical laboratory, fetal fraction, fetal sex, z-scores and/or a composite risk score for chromosomes 21/18/13, and the pregnancy outcome (e.g., diagnosis of trisomy 21, otherwise diploid for chromosomes 18 and 13). Upon receipt, samples were processed according the standard clinical protocols, aliquoted into 4-mL tubes and stored at −70 °C. Residual samples were tested at WIH for infectious diseases including hepatitis B surface antigen, anti-HCV, anti-HIV 1 and 2, and syphilis.
A “low risk” sample was defined as originating from a woman with no high risk factors (e.g., positive family history, positive serum screening test, abnormal ultrasound) and a negative clinical cfDNA test for chromosomes 21/18/13 (e.g., z-scores <2.0 and/or risk <1:10,000). Although these criteria do not exclude the possibility of an affected fetus being distributed as a “normal” challenge, the Down syndrome risk in these samples was estimated to be about 1 in 50,000 (1:500 [prevalence] × 0.5 [no high risk factors] * (1−0.98) [detection rate for cfDNA]). A “high risk” sample was defined as originating from women with both a positive clinical cfDNA test and a confirmatory diagnostic result. Exclusion criteria for all samples included a clinically reported fetal fraction <7%, positive for infectious disease, mosaic karyotype, and multiple gestations.
Participants
There were no geographical limitations on laboratory enrollment. The result form solicited information about the NIPS methodology, sequencing platform, and ancillary information such as the fetal fraction methodology and results. The laboratory’s clinical interpretation for chromosomes 21, 18, and 13 were also sought, as well as results for fetal sex. Not all enrolled laboratories reported results for both and six never returned any results. Throughout this document, laboratory and participant are synonymous terms.
Statistical analyses
The exercise data were extracted from the CAP Information Systems (Northfield, IL) and included all returned result forms, including those submitted after the closing date. Data were organized in an Excel spreadsheet (Microsoft, Redmond, OR) and analyses were performed in Excel or Prism v7.04 (La Jolla, CA). Graphics were produced in GraphPad/Prism (La Jolla, CA). Data from both distributions in 2018 were sometimes used to determine the appropriate classification, such as sequencing instrument manufacturer. Analyses were generally descriptive and included proportions, ranges of results, scatterplots, medians, averages, standard deviations, and 95% confidence intervals. When inferential analyses were employed, the test used as well as two-sided p values were reported. A significance level of 0.05 was used.
RESULTS
Participants and their locations
Overall, 66 subscriptions for the exercise were received. One laboratory ordered two subscriptions but only one set was included for analysis (randomly chosen). The 65 laboratories were located in Asia (26, 40%), North America (19, 29%), Europe (11, 17%), Africa (4, 6%), South America (3, 5%), and Australia/Oceania (2, 3%). Six laboratories (9%) failed to return any results (four from Asia and two from North America). Not returning results may be due to the sample condition upon receipt or other reasons and should not be taken as an indication of a test failure. Among the remaining 59 participants, 5 did not report cfDNA test results, possibly because they were in the process of implementing testing. This left a maximum of 54 responses for queries regarding NIPS results. However, some participants that did not return NIPS results did provide responses concerning testing platform and methodology as well as for selected supplemental questions.
Test information
Participants provided the sequencing platform manufacturer as well as their NIPS methodology. Table 1 shows the results for 56 respondents cross-tabulated for these two characteristics. Overall, 40 (69%) laboratories reported the manufacturer as Illumina (San Diego, CA) with the most common platform being the NextSeq (28/40, 70%). ThermoFisher (Waltham, MA) was next with 11 (19%), followed by Affymetrix (Santa Clara, CA) with 3 (5%) and BGI-Seq (Hong Kong, China) with 2 (3%). The most common methodology reported was shotgun sequencing (24, 43%) followed by genome sequencing (15, 27%), targeted SNPs (11, 20%), and targeted sequencing (6, 11%). Participants were also asked to include the test name that was provided to clients or on their website and these are listed as a footnote to Table 1.
Gestational and maternal ages
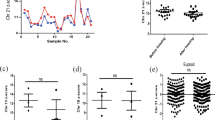
Laboratories must accurately determine the gestational age at sample collection to ensure it falls within their acceptable range for testing. The maternal history for each challenge included the date the ultrasound was performed and the gestational age at that time. Figure 1a shows the six challenges (horizontal axis) versus the reported decimal gestational age (vertical axis). For challenges 01, 02, 04, and 06, the sample draw date and the ultrasound study date were identical. Laboratories needed only to convert the provided days and weeks to decimal weeks and all provided correct responses (open circles). However, for challenges 03 and 05, the ultrasound was performed more than a week earlier than the sample drawn date and this interval must be added to obtain the correct gestational age. For challenge 03, 49% (18/37) correctly reported 13.3 weeks (open circles) while 51% (19/37) incorrectly reported 11.6 weeks (“X” symbol). For sample 05, a higher proportion (63%, 30/48) correctly reported 13.4 weeks while 37% (17/48) incorrectly reported 11.3 weeks. There was also one outlier. Seven of the 19 laboratories (37%) with incorrect responses for challenge 03 provided the correct gestational age for challenge 05.
College of American Pathologists (CAP) 2018 Educational Exercises: analysis of the reported decimal gestational age and decimal maternal age. (a) Noninvasive prenatal testing (NIPT) challenge is on the horizontal axis and the reported decimal gestational age is on the vertical axis. The data have been dithered left and right for visibility. For challenges 01, 02, 04, and 06, the date of the ultrasound was the same as the sample draw date and all participants provided correct responses. For samples 03 and 05, the ultrasound had been performed prior to the sample draw date. Those laboratories incorrectly reporting the lower gestational age (e.g., 11.6 weeks rather than 13.3 weeks for sample 03) are incorrect (indicated by X). Extreme values are also indicated by an X. (b) NIPT challenge is on the horizontal axis and the reported decimal maternal age is on the vertical axis. The decimal maternal age should be determined at the estimated date of delivery, several months later than the sample draw date. For all samples, a subset of laboratories incorrectly computed the maternal age. For sample 01, for example, the correct response was 25.7 years (higher set of observations, O) while the incorrect response was 25.3 years (lower set of observations, X). Any extreme results are also indicated by an X.
Laboratories also must be able to correctly compute the maternal age at the estimated date of delivery as tables and algorithms to compute the a priori Down syndrome risk are based on birth prevalences.18,19,20 Computing a woman’s age at sample draw date would underestimate her age at delivery and, subsequently, the fetal risks. Figure 1b shows the reported maternal ages. In distribution A, 69% (25/36) of respondents provided the appropriate maternal age at the estimated date of delivery. Those 11 participants reporting the incorrect maternal age are slightly lower on the plot (“X” symbol). Among these 11 participants, 10 reported results in distribution B and 6 (60%) had corrected their errors.
Fetal fraction
All 56 participants responding to whether they measure fetal fraction (percentage of total cfDNA derived from the fetoplacental unit) answered that they do. Overall, 30 (54%) are based on sequencing counts, 16 (29%) are SNP-based, 16 (29%) are based on fragment length, and 24 (43%) utilize Y-chromosome counts. Individually, 14 participants used only SNPs, 14 used only sequence counts, 2 used only Y-chromosome counts, and 1 used only fragment length. Among the remaining participants, 21 utilized two of these methods, 3 used three methods, and 1 used all four methods. No one reported using a methylation-based method or any other method.
Each participant’s fetal fraction estimate can be compared with the intended response based on the donor’s clinical NIPS results. This can be expressed as a ratio (participant/clinical), similar to a multiple of the median (MoM). If a participant’s result matches the intended response, the MoM would be 1.00. A participant’s median MoM can then be computed for all six challenges along with the range of MoM levels. Figure 2a shows the results of this analysis for each of the 54 participants, sorted from the lowest to the highest median MoM. The horizontal solid line shows the expected median MoM of 1.0 while the two dashed horizontal lines at 0.80 and 1.20 MoM defines a region where there is reasonable agreement. The overall median MoM was 0.99 (range of medians 0.54 to 1.59) and 38 (70%) of participants had medians indicating reasonable agreement. It was also possible to stratify these fetal fraction MoM levels by geographic location. The 44 international participants reported 199 fetal fraction estimates while the 15 US laboratories reported 79 with geometric mean MoM levels of 0.91 and 1.07, respectively (p < 0.001, t-test after logarithmic transformation, logarithmic SDs of 0.16 and 0.13, respectively). Each distribution was composed of multiple lots. A lot is a set of aliquots from three specific donors that were sent to multiple participants. Theoretically, those laboratories should provide similar fetal fraction results for each challenge within a given lot. Thus, it would be possible to compare each participant’s reported fetal fraction for each sample with its intended response. Over the six challenges, there were more than 40 lots, so analyzing each would result in high variability due to small sample sizes. However, many lots had essentially the same intended fetal fraction and could be combined. Figure 2b shows the results for distribution A and Fig. 2c shows the results for distribution B. The “X” symbols are drawn at the lot’s intended response (horizontal axis) versus the median of all participant responses receiving that lot (vertical axis). These symbols are expected to fall along the line of identity (Y = X). However, there are indications that the participant’s fetal fraction results and intended responses do not always agree. In Fig. 2c, for example, there were two lots with the same intended fetal fraction of 11%. Six participants that received one lot (indicated by open squares) all reported very low fetal fractions, with four of the six below 4%. The second lot with the same intended target had all participants reporting fetal fractions of 13% or higher filled diamonds. Thus, the intended response may be a reasonable expectation for most challenges, but should not be considered a gold standard.
College of American Pathologists (CAP) 2018 Educational Exercise: analyses of the reported fetal fraction results. (a) Each of the 56 participant’s fetal fraction estimates is expressed as a ratio to the intended response, similar to a multiple of the median (MoM). The open circles are the median fetal fraction expressed in MoM while the bars indicate the range of values. Results are sorted from low to high median value. Filled circles indicate participants whose summary results are below 0.8 or above 1.2 MoM. (b) Intended fetal fraction on the horizontal axis and the reported fetal fraction for distribution A. The large “X” symbols are drawn at the mean reported fetal fraction for all challenges having that intended fetal fraction. In theory, these should follow the line of identity (Y = X, dashed line). (c) Same as (b) except that it displays results from distribution B. In (c) the bold open squares and filled diamonds at the intended fetal fraction of 11% are from two separate lots and are discussed in “Results.” NIPT noninvasive prenatal testing.
Result interpretations for common trisomies and fetal sex
Table 2 provides summary information regarding the results for the common autosomal trisomies and fetal sex. The first column provides the distribution and sample challenge. The next three columns show whether the fetal fraction was reported by the laboratory to be acceptable (Yes) or unacceptable (No) for testing. In a few instances the laboratory may not have provided a response (Missing). The next three groups of four columns provide the proportion of chromosome 21, 18, and 13 results that were screen negative (Yes), screen positive (No), failed testing/no call (Failed) followed by the number of incorrect responses (Error). The last four columns in Table 2 summarize the reported fetal sex results for male or female fetuses, those not reporting results and the total number of errors. For the A distribution (challenges 01 to 03), no samples failed, all reported that the fetal fractions were acceptable and all trisomy classifications and fetal sex calls were correct. All 41 laboratories reported challenges 01 and 03 to be normal (diploid) for chromosomes 21, 18, and 13. All 40 laboratories receiving a trisomy 21 challenge for challenge 02 reported the sample to be abnormal (trisomic) for chromosome 21, and normal for chromosomes 18 and 13. One laboratory received a normal challenge and correctly classified it as being diploid for the three autosomes. Distribution B consisted of three nontrisomic challenges and results from all successful tests were correct, including the fetal sex calls. However, among the 143 tests performed there were 10 test failures (7.0% in B, 3.7% overall), half of which were likely due to samples having relatively low fetal fractions measurements.
Laboratories reporting underlying z-scores
In the A distribution, the result form did not allow for negative z-scores, so results could not be analyzed for the two euploid challenges. However, it was assumed that all z-scores for challenge 02 (confirmed trisomy 21) were positive since all laboratories reported that sample to be screen positive. One laboratory consistently reported extremely high z-scores for all three chromosomes, regardless of aneuploidy status and another received a normal challenge. These two results were excluded. Of the remaining 21 results, the mean chromosome 21 z-score was 12.3 (median 12.3, range 3.6 to 26.7). Figure 3 shows these z-scores versus the reported fetal fraction. Filled symbols represent US laboratories while open symbols represent international laboratories. Each symbol represents a different lot. Responses from US laboratories are somewhat higher but are associated with lots having higher fetal fractions. The regression line shows the well-established positive relationship between fetal fraction and z-score.5 One exception is the observation with the lowest z-score (3.6) that had a relatively high fetal fraction (16.8%). Another laboratory receiving the same lot reported the same fetal fraction but a much higher z-score of 25.2. All other z-scores were 5.0 or higher and all reported fetal fractions were 6% or higher. No information regarding interim results were reported by the remaining participants.
Sample challenge NIPT-02: reported z-scores and fetal fractions from 21 participants in the educational exercise. These sample lots were collected from women with a confirmed fetus with trisomy 21 (Down syndrome). For each laboratory, the reported fetal fraction is shown on the horizontal scale and the reported chromosome z-score on the vertical axis. The vertical line at a 4% fetal fraction indicates a level at which some laboratories will consider the level to be too low for reliable testing. The horizontal line at a z-score of 3 is a commonly used cut-off to classify results as screen positive (at or above 3) or screen negative (below 3) for Down syndrome. Open symbols show responses from international laboratories and filled symbols show responses from laboratories located in the US. Each symbol indicates a different lot, where those laboratories received an aliquot from the same enrolled pregnancy. All successfully identified this sample from a fetus affected with Down syndrome (trisomy 21). The regression lines show the known relationship of increasing z-scores with increasing fetal fraction, but the slope for the 21 observations is not statistically significant (p = 0.18). The z-scores are generally higher for the US laboratories but so are the corresponding intended fetal fractions.
In the B distribution, the intended responses were all diploid (normal) for chromosomes 21, 18, and 13. A total of 33 laboratories reported usable z-scores. For challenge 04, the mean z-scores were −0.4 (range −3.4 to 1.9), −0.3 (−2.3 to 1.3), and −0.5 (−3.4 to 1.4). For challenge 05, these results were −0.5 (range −4.9 to 2.2), 0.0 (−1.5 to 2.8), and −0.6 (−2.6 to 2.5). For challenge 06 these results were −0.2 (range −7.2 to 2.4), 0.0 (−2.1 to 2.3), and −0.4 (−2.7 to 1.1).
Responses to supplemental questions
In both the A and B distributions, supplemental questions were included in the exercise. Information about turnaround time was reported by 52 participants. The median was 7 days (range 3 to 21 days). Of these, 19 (37%) reported 5 days or less while 13 (25%) reported 10 days or more. The 38 international participants had a median of 7 days (range 3 to 21), while 13 US laboratories had a median of 5 days (range 3 to 9). Unfortunately, the definition of turnaround time is not standardized. Some define it as sample receipt at the laboratory to the time results are available, others count from the draw date to the report date. Also, some count only working days while others count all intervening days, making comparisons difficult.
The actual text regarding the description of a screen positive test result for chromosome 21 was reported by 43 participants. Most were unique. Thus, a scoring system was developed using the following: inclusion of the term “Down syndrome” was worth two points, the term “trisomy 21” was worth one point. Term(s) such as “risk,” “screen positive,” or “probability” were worth two points. If none of these “risk-related” terms were included, the score was set to 0 with the reasoning that the result could be misinterpreted as being diagnostic. Using this system, seven participants (16%) included all intended information with responses such as “High risk for trisomy 21 (Down syndrome).” Another 16 (37%) scored 0, often because of seemingly diagnostic responses such as “Aneuploidy detected consistent with trisomy 21.” Another 20 participants (47%) scored 3, usually by leaving out the term “Down syndrome.” The average score was 2.2 and this did not differ when stratified by location (11 US participants = 2.0, 32 international participants = 2.3; exact Mann–Whitney, p = 0.75).
Clinical validity information included on patient reports was provided by 42 participants. The detection rate (sensitivity) was included for 34 (81%) while the false positive rate (specificity) was included in 26 (62%). The positive predictive value was reported by 14 (33%) and the negative predictive values by 5 (11%). Fourteen (33%) reported a patient-specific risk and 2 (5%) reported an alternative measure: accuracy. Accuracy is the total number of correct calls (true positive + true negative) divided by the numbers tested and does not distinguish between detection and false positive rates. These responses can be compared with a recent similar survey of laboratories located in the United States.21
Epidemiological monitoring done as part of routine quality assessment was reported by 43 laboratories. The screen positive rate was monitored by 40 participants (93%), 40 (93%) monitor the test failure/no call rate, 29 (67%) monitor the repeat sampling rate (requested and/or received), and 38 (88%) monitor the rate of low fetal fraction. Whether a sample is drawn too early was monitored by 37 (63%).
Additional supplemental questions included the range of acceptable gestational ages, types and rates of test failures, actions taken when a test failure/no call is reported, testing the sex chromosomes, and use of maternal weight. Summaries of responses to these queries can be found in the Supplemental Materials.
DISCUSSION
The 2018 CAP NIPT Educational Exercises demonstrate that patient-derived plasma works well for the multiple methods, platforms, and laboratory developed protocols represented by participants offering NIPS around the world. Among the 266 clinical calls provided by participants (123 in distribution A and 143 in distribution B), there were no genotyping errors for chromosomes 21, 18, and 13. This compares favorably with the 2.3% error rate reported in the European program,16 but that survey did include more trisomic challenges.16 Among the 214 fetal sex calls (96 in A and 118 in B), all were correct. However, one laboratory did report a male sample as being screen positive for 47,XXY. The reported fetal fraction generally agreed with the intended response, but there may be issues with consistently lower than expected fetal fractions, at least for one lot (identified in Fig. 2c). No test failures/no calls occurred in distribution A (0 of 123), but 10 occurred in 143 responses in distribution B. Overall, the test failure rate was 3.6% (10/276), similar to the 2.3% reported in the European report.16 However, three of these failures can be attributed to a sample with a fetal fraction that was likely below the generally accepted threshold of 4%. Among the remaining seven failures, two were associated with relatively low fetal fractions (4.7, 5.6) but the laboratories reported them to be acceptable. The intended fetal fraction of the challenges, as reported on a clinical NIPS report, was always 7% or higher. However, the fetal fraction estimates can have up to a 20% coefficient of variation and setting a lower limit of 7% provides some cushion against distributing challenges that fall close to or below 4%. There was a single lot for which all of the laboratories determined the fetal fraction to be much lower than the intended response (Fig. 2c) and the reason for this is not clear.
These data also provided evidence that an important percentage of laboratories were unable to correctly compute the gestational age and/or the maternal age. Both of these are important characteristics for determining whether the test should be performed, and in computing the age-related risks. The maternal age discrepancies are likely due to many of these laboratories being focused on molecular diagnostics with limited experience in prenatal screening as it is well known that the age at delivery is needed. The same reasoning is likely responsible for the gestational age discrepancies as well. However, one mitigating factor is that gestational age is not as important in interpreting cfDNA test results as it is in interpreting serum screening results. Regardless, the correct computation of gestational age at the sample draw is still important in determining whether the sample is too early for reliable cfDNA testing. Importantly, 37% and 60% of laboratories informed of their miscalculations in the A distribution for maternal age and/or gestational age, corrected responses in the B distribution, indicating that participating in these exercises can improve clinical laboratory practice.
For those laboratories that report interim results as z-scores (or normalized chromosome values) the results show the expected positive association with fetal fraction. One laboratory, however, reported a very high fetal fraction and a z-score that was close to the lower cut-off level. This is worrisome, as that laboratory may miss trisomic pregnancies when the fetal fraction is considerably lower, but still acceptable.
In general, the language used to describe the screening results is not consistent between laboratories or easily interpreted. There are opportunities for misinterpretation by primary care providers that the test is diagnostic (e.g., “Aneuploidy detected”). Given the decades-long history of prenatal screening for these common trisomies, we recommend that laboratories continue using the phrase “Screen positive for…” and “Screen negative for….” This reinforces that the test is not diagnostic, but coupled with the word “positive” informs both the health-care provider and patient with a key to test interpretation.
The results of this exercise have limitations. It is difficult to obtain sufficient numbers of usable specimens, especially for trisomic pregnancies. Thus, we were only able to distribute one Down syndrome sample out of six challenges. This issue is being addressed and we hope to have a wider array of disorders represented more often in the future. A recent publication22 described a method of manufacturing proficiency samples that may be suitable for a wide range of methodologies, but more extensive validation is needed.
In summary, the use of real patient samples is suitable for all NIPS methods and technologies represented by participating laboratories and there were no genotyping errors for chromosomes 21, 18, and 13. All fetal sex calls were also correct. Among the test failures that did occur, up to half were likely due to low fetal fraction while the remaining failures were for other reasons. We also observed that feedback from the first distribution led to correction of computational errors in the second distribution, indicating that laboratories are reacting seriously to the exercise’s results as a way to help improve the quality of NIPS testing and reporting. Although specific laboratory results for this exercise are not available to the public, individuals can request information from any laboratory concerning their method for demonstrating external quality assessment and for any results that might be available. However, laboratories have no legal obligation to respond. An exercise such as the one described here is only one of many factors to consider when choosing a NIPS laboratory for clinical testing. The findings presented here provide some level of confidence in the clinical validity of the NIPS results generated from a wide range of methods used by laboratories throughout the world.
References
Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487.
Devaney SA, Palomaki GE, Scott JA, Bianchi DW. Noninvasive fetal sex determination using cell-free fetal DNA: a systematic review and meta-analysis. JAMA. 2011;306:627–636.
Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–16271.
Chiu RW, Chan KC, Gao Y, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:20458–20463.
Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13:913–920.
Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2015;45:249–266.
Korpi-Steiner N, Chiu RWK, Chandrasekharan S, et al. Emerging considerations for noninvasive prenatal testing. Clin Chem. 2017;63:946–953.
Minear MA, Alessi S, Allyse M, Michie M, Chandrasekharan S. Noninvasive prenatal genetic testing: current and emerging ethical, legal, and social issues. Annu Rev Genomics Hum Genet. 2015;16:369–398.
Kotsopoulou I, Tsoplou P, Mavrommatis K, Kroupis C. Non-invasive prenatal testing (NIPT): limitations on the way to become diagnosis. Diagnosis (Berl). 2015;2:141–158.
Pergament D, Ilijic K. The legal past, present and future of prenatal genetic testing: professional liability and other legal challenges affecting patient access to services. J Clin Med. 2014;3:1437–1465.
Gregg AR, Gross SJ, Best RG, et al. ACMG statement on noninvasive prenatal screening for fetal aneuploidy. Genet Med. 2013;15:395–398.
Takoudes T, Hamar B. Performance of noninvasive prenatal testing when fetal cell-free DNA is absent. Ultrasound Obstet Gynecol. 2015;45:112.
Palomaki GE, Ashwood ER, Weck KE. A flawed challenge but valid recommendation: a response to Takoudes and Hamar. Ultrasound Obstet Gynecol. 2015;45:117.
UK NEQAS. https://ukneqas.org.uk/. Accessed 7 October 2019.
EMQN. https://www.emqn.org/. Accessed 7 October 2019.
Deans ZC, Allen S, Jenkins L, et al. Ensuring high standards for the delivery of NIPT world-wide: Development of an international external quality assessment scheme. Prenat Diagn. 2019;39:379–387.
College of American Pathologists. Non-Invasive Prenatal Testing - NIPT. https://estore.cap.org/OA_HTML/xxCAPibeCCtpItmDspRte.jsp?section=10469&item=567578&sitex=10020:22372:US. Accessed 7 October 2019.
Hecht CA, Hook EB. The imprecision in rates of Down syndrome by 1-year maternal age intervals: a critical analysis of rates used in biochemical screening. Prenat Diagn. 1994;14:729–738.
Cuckle HS, Wald NJ, Thompson SG. Estimating a woman's risk of having a pregnancy associated with Down's syndrome using her age and serum alpha-fetoprotein level. Br J Obstet Gynaecol. 1987;94:387–402.
Morris JK, Mutton DE, Alberman E. Revised estimates of the maternal age specific live birth prevalence of Down's syndrome. J Med Screen. 2002;9:2–6.
Skotko BG, Allyse MA, Bajaj K, et al. Adherence of cell-free DNA noninvasive prenatal screens to ACMG recommendations. Genet Med. 2019;21:2285–2292.
Zhang R, Ding J, Gao P, Li Z, Tan P, Li J. Generation of highly biomimetic quality control materials for noninvasive prenatal testing based on enzymatic digestion of matched mother-child cell lines. Clin Chem. 2019;65:761–770.
Acknowledgements
We thank Geralyn M. Lambert-Messerlian, Edward M. Kloza, and Elizabeth Eklund of the Division of Medical Screening and Special Testing at Women & Infants Hospital of Rhode Island for their work in organizing maternal blood sample collection, processing, data collection, storage, and labeling of aliquots for shipment to laboratories participating in these educational exercises. We thank the prenatal care providers who approached women and obtained the blood samples needed for these exercises as well as the women who donated blood. We also thank Pamela Provax, Technical Manager, College of American Pathologists, for her support of the NIPT Educational Exercises concept and implementation. Lastly, we thank those laboratories who participated in the multiple pilot trials that provided invaluable baseline information for our work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
All authors are members of the Biochemical and Molecular Genetics (BMG) Committee of the College of American Pathologists. G.E.P. is a member of the Division of Medical Screening and Special Testing at Women & Infants Hospital (WIH) that holds the current contract with the College of American Pathologists to manufacturer the samples used for the NIPT Educational Exercises. He is also the principal investigator (PI) of an industry-sponsored study at WIH to assess the clinical validation of a non-PCR-based method for NIPS (ClinicalTrials.gov identifier: NCT03087357) and was PI on previous similar studies (NCT00877292 and NCT01966991). Through his employer, he has consulted for Ansh Labs (Webster, TX), Illumina (San Diego, CA), Roche (Indianapolis, IN), PerkinElmer (Waltham, MA), and LabCorp (Burlington, NC).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Palomaki, G.E., Wyatt, P., Best, R.G. et al. Assessment of laboratories offering cell-free (cf) DNA screening for Down syndrome: results of the 2018 College of American Pathology External Educational Exercises. Genet Med 22, 777–784 (2020). https://doi.org/10.1038/s41436-019-0718-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-019-0718-4