Abstract
Purpose
Noninvasive prenatal screening (NIPS) using genome sequencing also reveals maternal copy-number variations (CNVs). Those CNVs can be clinically actionable or harmful to the fetus if inherited. CNVs in the DMD gene potentially causing dystrophinopathies are among the most commonly observed maternal CNVs. We present our experience with maternal DMD gene CNVs detected by NIPS.
Methods
We analyzed the data of maternal CNVs detected in the DMD gene revealed by NIPS.
Results
Of 26,123 NIPS analyses, 16 maternal CNVs in the DMD gene were detected (1/1632 pregnant women). Variant classification regarding pathogenicity and phenotypic severity was based on public databases, segregation analysis in the family, and prediction of the effect on the reading frame. Ten CNVs were classified as pathogenic, four as benign, and two remained unclassified.
Conclusion
NIPS leverages CNV screening in the general population of pregnant women. We implemented a strategy for the interpretation and the return of maternal CNVs in the DMD gene detected by NIPS.
Similar content being viewed by others
INTRODUCTION
Noninvasive prenatal screening (NIPS) based on the analysis of cell-free DNA (cfDNA) in the maternal serum has enabled the detection of fetal aneuploidies, typically trisomy 21, 13, and 18, with a high sensitivity and specificity.1,2 Because of its high accuracy, NIPS was rapidly implemented as part of routine prenatal care. Notably, genome massively parallel sequencing allows not only the analysis of the viable trisomies, but also fetal aneuploidies of other chromosomes, and has the potential to detect smaller fetal chromosomal imbalances as well.3,4,5 Moreover, since on average 90% of the cfDNA in the maternal circulation originates from maternal cells, a genome-wide cfDNA analysis equally results in a high resolution screen for maternal copy-number variations (CNVs).6 Hence, NIPS has the potential to detect relevant maternal CNVs that can be clinically actionable or could be potentially harmful for the fetus if inherited.6,7,8
Duchenne muscular dystrophy (DMD) is an X-linked condition that presents as a progressive muscle disorder in young boys, affecting both skeletal and cardiac muscle.9 With an incidence at birth of 1/3800 to 1/6300, it is one of the most common severe, untreatable neuromuscular disorders.10 It is caused by a loss-of-function variant in the DMD gene (Xp21.2-p21.1), and represents the severe end of the spectrum of diseases collectively called dystrophinopathies. Becker muscular dystrophy (BMD), in contrast, is a milder dystrophinopathy, with onset later in childhood and with an incidence of 1 in 20,000 to 30,000 males. A milder phenotype is DMD-associated cardiomyopathy, also called X-linked dilated cardiomyopathy, with onset usually in young adults.11 Asymptomatic individuals with isolated elevated serum creatine kinase (CK) have also been reported. Most carrier females are asymptomatic, but there is an increased risk for muscle damage and dilated cardiomyopathy.12 Of interest, the DMD gene is one of the largest genes in the human genome, with 79 exons, and most of its variants correspond to deletions (60–70%) and duplications (5–10%), the remaining being single-nucleotide variants. Given that a large proportion of variants in the DMD gene are inherited CNVs, genome-wide NIPS is expected to detect carrier mothers for DMD.9
When a CNV in the DMD gene is detected, sometimes it is challenging to predict the phenotype for male carriers. The most reliable phenotypic predictions are based on family history. However, in population-based genome-wide analysis, family histories of dystrophinopathy are lacking. To interpret the potential pathogenicity, different databases with genotype–phenotype correlations of DMD gene variants can be consulted. The best known are the Leiden Open Variation Database (LOVD) and the UMD-DMD France Database.13,14 The effect of novel hitherto unreported CNVs may be predicted using the frameshift rule, which permits to distinguish DMD from BMD with a 91–92% accuracy.9 In general, frameshift variants cause DMD whereas in-frame variants cause BMD.13 However, exceptions exist, e.g., 2% of out-of-frame deletions and duplications will actually result in BMD and 7% of all DMD cases are caused by an in-frame variant. This indicates that other factors including size and position of the CNV may also play a role. The interpretation is further complicated by the variable expression observed for certain CNVs with apparently the same breakpoints.13 Whether or not to report a maternal CNV detected by NIPS depends on reliable prediction of the phenotypic consequences of that specific CNV.
The Belgian Advisory Committee on Bioethics stated that secondary findings of clinical relevance detected during NIPS should be reported: “Where this information may lead to a preventive or therapeutic intervention, it is important to also share this information with the patient in the context of clinical genetics. The failure to do so may be construed as serious negligence.”15 This prompted the Belgian Society for Human Genetics to issue guidelines on how to deal with clinically significant secondary findings, both maternal and fetal, detected by NIPS.16 According to these guidelines, carriership for X‐linked recessive disorders will be communicated, irrespective of the sex of the fetus. As of 1 July 2017, NIPS is reimbursed by the National Health System for all pregnant women in Belgium. In this study, we report a systematic review of the cases where NIPS revealed a maternal CNV involving the DMD gene from a single genetic center, during a 1-year period following the universal reimbursement in Belgium. Since current expertise on this topic is limited, and guidelines are either lacking or based on expert opinion, here we propose a strategy to interpret and return variants in the DMD gene.
MATERIALS AND METHODS
Sample collection
This study was approved by the Ethical Committee at UZ/KU Leuven (MP004717). Written informed consent for returning secondary findings was obtained from all the participants by the referring physician. The study population consists of all pregnant women in Belgium undergoing a NIPS at the Center for Human Genetics, UZ Leuven, Leuven, Belgium over a 1-year period from 1 July 2017 to 30 June 2018.
Genetic technology
NIPS and CNV detection were carried out as described.3,6,17 Briefly, low-pass genome sequencing generated ~10 million single-end reads of 36 bp per sample, corresponding to a 0.1× coverage. These reads were mapped to human genome build 19 (hg19), and polymerase chain reaction (PCR) duplicates were removed. The resulting genomic coordinates of read starts served as input to CNV caller SeqCBS,21 in combination with a sex-matched control obtained by pooling read start coordinates from 50 male (46,XY) or 50 female (46,XX) “normal” pregnancies. To confirm the presence of the respective maternal CNVs by chromosomal microarray analysis, DNA was extracted from maternal white blood cells, obtained from the stored buffy coats of the NIPS blood sample. Array comparative genomic hybridization (array-CGH) was performed using the 8 × 60 K CytoSure ISCA v3 microarray (Oxford Gene Technology Oxford, UK) as described.18 Array-CGH genomic coordinates are based on human genome build 19 (hg19).
Variant classification
The data were retrieved from patient and laboratory files. Phenotype predictions were based on the following principles. First, we assessed whether the CNVs had already been described in patients with a dystrophinopathy, using two publicly available databases. The UMD-DMD France database describes both published and unpublished genetic and clinical information about variants in the DMD gene. Molecular data were derived from 14 diagnostic laboratories in France. The database only includes CNVs with known exon boundaries. The Leiden Open Variation Database (LOVD) contains genetic and in many cases also phenotypic data from over 16,000 patients with a DMD gene variant. Second, we determined whether the CNV was predicted to be in-frame or out-of-frame, using the online tool in the LOVD. Third, for the reported variants, segregation analysis within the family was proposed during the counseling, using as many available and potentially informative individuals as possible. Clinical information included a detailed history and, when available, serum creatine kinase values.
RESULTS
Maternal CNVs in the DMD gene
Sixteen maternal CNVs were detected in the DMD gene of 26,123 NIPS analyses performed. This corresponds to an incidence of 1/1632 pregnant women. The data are summarized in Table 1.
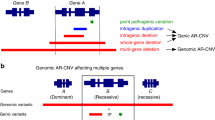
The genomic representation of the region containing the DMD gene, displaying the 16 maternal CNVs detected by NIPS analysis, is illustrated in Fig. 1. At 0.1× coverage, the performance of SeqCBS CNV calling can be expected to vary per CNV size and per region. Therefore, confirmation with an independent technique is necessary, especially for smaller CNVs and if the exonic content of a CNV determines the clinical outcome, as is the case for DMD/BMD. The size and position of all CNVs were validated by chromosome microarray analysis, using DNA extracted from maternal white blood cells (Table 2). The array platform includes probes for up to 502 highly targeted developmental delay genes enabling detection of single-exon aberrations. Higher probe density across the exons of the DMD gene allows improved detection of exonic CNVs. All 16 maternal CNVs detected by NIPS were confirmed by chromosomal microarray analysis. In four cases, the array-CGH further refined the CNV size and exonic content (families 4, 6, 8, and 12) (Table 2). Figure 2 shows an example (family 4) of array-CGH refinement of one DMD CNV detected by NIPS.
Genomic representation of the Xp21.2-p21.1 region encompassing the DMD gene. University of California–Santa Cruz (UCSC) genomic representation of the X chromosome encompassing the DMD gene at Xp21.2-p21.1 displaying the 16 maternal CNVs detected by NIPS. The duplications and deletions are represented as blue and red bars, respectively. CNVs copy-number variations; NIPS noninvasive prenatal screening.
Example of one DMD CNV detected by NIPS. (a) Ideogram of chromosome X highlighting the Xp21.2-p21.1 region encompassing the DMD gene. (b) NIPS CNV result, represented as red bar, with the corresponding exons tracks included in the DMD CNV call. (c) Array-comparative genomic hybridization (array-CGH) result showing the refinement of DMD CNV size. Image of array-CGH data was extracted from CytoSureTM Interpret software. CNV copy-number variation; NIPS noninvasive prenatal screening.
Variant classification
Of the 16 families, 3 carried the same recurrent in-frame duplication of exons 10–27 (families 10, 11, and 12). Thus, in total, 14 different CNVs were observed. Nine of these 14 CNVs (families 1–9) were present in the databases with sufficient phenotypic data, enabling correct interpretation. In addition, family segregation data were available in five of these nine families (families 1–5). In three families (families 1–3), the CNV had previously been diagnosed. In family 1, the mother has a son affected with DMD and was a known carrier of a pathogenic DMD deletion (exon 51). The pregnancy was established after preimplantation genetic diagnosis (PGD). In family 2, the CNV was detected as a secondary finding by microarray-CGH in her nephew with unexplained intellectual disability. This information was not known to us when the NIPS was performed. In family 3, segregation confirmed the presence of a mild BMD phenotype described in the literature. In two families (families 4 and 5) segregation was not informative; in family 4 the variant was maternally inherited, and in family 5 the variant is de novo so no clinical information was gained. In the remaining four families (6–9) no segregation analysis could be performed.
The five remaining CNVs (families 10–16) had not been reported before and further steps were needed to classify them. Segregation analysis enabled classification of two. Three apparently unrelated women (families 10–12) carried a recurrent in-frame duplication of exons 10–27. The presence of this CNV in some clinically unaffected adult male relatives allowed classification of the CNV as likely benign. Therefore, this variant is considered as a rare local benign CNV. Also in family 13, paternal inheritance allowed classification of this CNV as likely benign. One novel out-of-frame duplication (family 14, exons 51–62), for which family segregation was not informative, was classified as likely pathogenic for DMD. The variants detected in family 15 and family 16 remained unclassified.
Based on the experience we present here, we propose a strategy for the interpretation and reporting of CNVs in the DMD gene (Fig. 3).
DISCUSSION
Here, we report our experience with NIPS and the incidental detection of maternal CNVs in the DMD gene over a 1-year period. Because NIPS was reimbursed for all pregnant women in the time period studied, this population is likely to be representative of the Belgian population of reproductive women, with a standard risk of having a child with a dystrophinopathy. In a 1-year period, we performed 26,123 NIPS tests and we observed 16 CNVs in the DMD gene. CNVs in DMD are thus present in 1/1632 women.
Variant classification is challenging, especially in a setting where the DMD CNV is not ascertained through an affected family member. Nguyen et al. reported chromosomal microarray data of five women referred for diagnosis of intellectual disability with a CNV in the DMD gene.19 In two, the CNV was paternally inherited and classified as benign, but in the other three, the CNV was not detected in any male family member. This led to challenges regarding disclosure of genetic risk to the family. Of the 16 families investigated in this study, 3 carried a recurrent in-frame duplication of exons 10–27. Thus, 14 different CNVs were observed. The most reliable predictor of the phenotype relies on previously known genotype–phenotype correlations, either from affected family members or from the databases. Of interest, only 9 of the 14 CNVs have been reported in public databases. Of these nine, four were associated with likely or known DMD, two with likely BMD, and three with a variable phenotype. Segregation analysis in the family can contribute to variant classification. In two families the mother was a carrier of a previously detected familial DMD CNV: (1) a pathogenic out-of-frame DMD deletion (exon 51), resulting in the expected Duchenne muscular dystrophy phenotype in her son; and (2) an in-frame duplication without associated phenotypic manifestations in this family. Segregation could be carried out in three families: in two families it was not informative and in one it confirmed the mild BMD phenotype described in the literature. Of the five novel variants, segregation analysis could be performed for three. Based on this analysis two CNVs were classified as likely benign while one family was not informative. The reading frame rule predicts that out-of-frame CNVs have a more than 90% risk of being associated with DMD. The out-of-frame duplication of exons 51–62 was therefore reclassified from variant of unknown significance (VUS) to likely pathogenic. A major challenge in variant classification is the observation that for three of the detected CNVs, the databases report a variable phenotype. Variability in expression is well known in DMD gene variants. Several explanations have been put forward. One possibility is that the positions of the CNVs are not exact, due to an imperfect resolution of the technologies used.13,20,21 In a diagnostic setting, the phenotype is usually clinically evident and relies less on the underlying genetic defect. In contrast, imperfect breakpoint mapping may hamper the prediction of the phenotype in a family without affected individuals.
After exclusion of the two previously known carrier women, of the 16 pregnancies, 8 carried a CNV associated with a dystrophinopathy, conferring a 50% risk of an affected male fetus in 6 of them. From the incomplete follow-up data, we know that for two male fetuses at risk for DMD, an invasive test was performed, with one normal result and one affected result. However, even in pregnancies with a female fetus, this information can be important for the mother herself, given the increased risk of manifestations for future pregnancies and for relatives. In accordance with the guidelines from the Belgian Society for Human Genetics on secondary findings detected by NIPS, these results were reported.
One of the main challenges in managing secondary findings during prenatal diagnosis is maintaining the balance between providing actionable genetic information and not returning information that harms the patient.20 The actionability of returning CNVs with known or high risk of being associated with DMD is evident. However, this is less evident for BMD-associated variants. This is further complicated by the fact that the severity of BMD can be highly variable. Therefore, segregation analysis can aid in variant classification, as it did in some of the families in this study. However, this necessitates reporting and discussing the results, which may cause unnecessary anxiety. There are several arguments against reporting unclassified variants, unless they are associated with a high risk of a severe phenotype, i.e., when they cause a frameshift. First, novel CNVs, absent in the very extensive disease databases, are more likely to represent benign CNVs without phenotypic consequences. Second, segregation analysis is unlikely to provide evidence for severe phenotypic consequences, but rather for a likely benign effect when observed in unaffected male adults. Classification as DMD- or BMD-associated can only be achieved in the case of a positive family history for the disease. A preconception family history should have revealed this. Third, after reporting the variants, genetic counseling was offered to all couples without any delay, together with physicians from the neuromuscular team when indicated. In one of the pregnancies where a novel CNV was found, the couple decided to terminate the pregnancy after their healthy 3-year-old son was found with mildly elevated CK. They opted not to wait for the results of the healthy maternal father, who was subsequently also found to be a carrier.
In summary, NIPS results can return relevant and clinically actionable variants to the mother. We show that NIPS leverages a screening in the general population of pregnant women, which enables detection of CNVs causing dystrophinopathies. Because no studies exist on CNVs in the DMD gene in women following NIPS, and current expertise on this topic is limited, we present a strategy to interpret and return variants in the DMD gene.
References
Bianchi DW, Chiu RWK. Sequencing of circulating cell-free DNA during pregnancy. N Engl J Med. 2018;379:464–473.
Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, Das AF, et al. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370:799–808.
Brison N, Neofytou M, Dehaspe L, Bayindir B, Van Den Bogaert K, Dardour L, et al. Predicting fetoplacental chromosomal mosaicism during noninvasive prenatal testing. Prenat Diagn. 2018;38:258–266.
Lefkowitz RB, Tynan JA, Liu T, Wu Y, Mazloom AR, Almasri E, et al. Clinical validation of a noninvasive prenatal test for genome-wide detection of fetal copy number variants. Am J Obstet Gynecol. 2016;215:227.e1–227.e16.
Li R, Wan J, Zhang Y, Fu F, Ou Y, Jing X, et al. Detection of fetal copy number variants by noninvasive prenatal testing for common aneuploidies. Ultrasound Obstet Gynecol. 2016;47:53–57.
Brison N, Van Den Bogaert K, Dehaspe L, et al. Accuracy and clinical value of maternal incidental findings during noninvasive prenatal testing for fetal aneuploidies. Genet Med. 2017;19:306–313.
Amant F, Verheecke M, Wlodarska I, Dehaspe L, Brady P, Brison N, et al. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol. 2015;1:814–819.
Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314:162–169.
Darras BT, Urion DK, Ghosh PS. Dystrophinopathies. 2000 Sep 5 [updated 2018]. In: Adam MP, Ardinger HH, Pagon RA, et al. GeneReviews® [Internet]. Seattle, WA: University of Washington, Seattle; 1993–2018. http://www.ncbi.nlm.nih.gov/books/NBK1119/.
Landfeldt E, Lindgren P, Bell C, et al. The burden of Duchenne muscular dystrophy: an international, cross-sectional study. Neurology. 2014;83:529–536.
Ferlini Sewry C, Melis MA, Mateddu A, Muntoni F. X-linked dilated cardiomyopathy and the dystrophin gene. Neuromuscul Disord. 1999;9:339–346.
Ishizaki M, Kobayashi M, Adachi K, et al. Female dystrophinopathy: review of current literature. Neuromuscul Disord. 2018;28:572–581.
Aartsma-Rus A, Van Deutekom JC, Fokkema IF, et al. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144.
Tuffery-Giraud S, Béroud C, Leturcq F, et al. Genotype-phenotype analysis in 2405 patients with a dystrophinopathy using the UMD-DMD database. A model of nationwide knowledgebase. Hum Mutat. 2009;30:934–945.
Belgian Advisory Committee on Bioethics. Opinion no. 66—noninvasive prenatal testing (NIPT). https://www.health.belgium.be/sites/default/files/uploads/fields/fpshealth_theme_file/opinion_66_web_1.pdf. Accessed 9 May 2016.
Belgian Society for Human Genetics. Belgian guidelines for managing incidental findings detected by NIPT. 2017. http://www.beshg.be/index.php?page=guidelines.
Bayindir B, Dehaspe L, Brison N, et al. Noninvasive prenatal testing using a novel analysis pipeline to screen for all autosomal fetal aneuploidies improves pregnancy management. Eur J Hum Genet. 2015;23:1286–1293.
Brady P, Brison N, Van Den Bogaert K, et al. Clinical implementation of NIPT—technical and biological challenges. Clin Genet. 2016;89:523–530.
Nguyen K, Putoux A, Busa T, et al. Incidental findings on array comparative genomic hybridization: detection of carrier females of dystrophinopathy without any family history. Clin Genet. 2015;87:488–491.
Aartsma-Rus A, Ginjaar IB, Bushby K. The importance of genetic diagnosis for Duchenne muscular dystrophy. J Med Genet. 2016;53:145–151.
Shen JJ, Zhang NR. Change-point model on nonhomogeneous Poisson processes with application in copy number profiling by next-generation DNA sequencing. Ann Appl Stat. 2012;6:476–496.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Brison, N., Storms, J., Villela, D. et al. Maternal copy-number variations in the DMD gene as secondary findings in noninvasive prenatal screening. Genet Med 21, 2774–2780 (2019). https://doi.org/10.1038/s41436-019-0564-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-019-0564-4
Keywords
This article is cited by
-
Population screening for 15q11-q13 duplications: corroboration of the difference in impact between maternally and paternally inherited alleles
European Journal of Human Genetics (2024)
-
X-CNV: genome-wide prediction of the pathogenicity of copy number variations
Genome Medicine (2021)
-
Duchenne muscular dystrophy
Nature Reviews Disease Primers (2021)
-
Outcome of publicly funded nationwide first-tier noninvasive prenatal screening
Genetics in Medicine (2021)