Abstract
Purpose
Multiple chromosomal aneuploidies may be associated with maternal malignancies and can cause failure of noninvasive prenatal screening (NIPS) tests. However, multiple chromosomal aneuploidies show poor specificity and selectivity for diagnosing maternal malignancies.
Methods
This multicenter retrospective analysis evaluated 639 pregnant women who tested positive for multiple chromosomal aneuploidies on initial NIPS test between January 2016 and December 2017. Women were assessed using genome profiling of copy-number variations, which was translated to cancer risk using a novel bioinformatics algorithm called the cancer detection pipeline (CDP). Sensitivity, specificity, and positive predictive value (PPV) of diagnosing maternal malignancies were compared for multiple chromosomal aneuploidies, the CDP model, and the combination of CDP and plasma tumor markers.
Results
Of the 639 subjects, 41 maternal malignant cancer cases were diagnosed. Multiple chromosomal aneuploidies predicted maternal malignancies with a PPV of 7.6%. Application of the CDP algorithm to women with multiple chromosomal aneuploidies allowed 34 of the 41 (83%) cancer cases to be identified, while excluding 422 of 501 (84.2%) of the false positive cases. Combining the CDP with plasma tumor marker testing gave PPV of 75.0%.
Conclusion
The CDP algorithm can diagnose occult maternal malignancies with a reasonable PPV in multiple chromosomal aneuploidies–positive pregnant women in NIPS tests. This performance can be further improved by incorporating findings for plasma tumor markers.
Similar content being viewed by others
INTRODUCTION
Noninvasive prenatal screening (NIPS) tests first became commercially available in 2011 to screen for fetal trisomy 21, and they globalized rapidly.1 Current NIPS tests can detect fetal trisomy 21, 18, and 13 with high sensitivity and specificity.2,3 However, NIPS tests can give false positive results or fail entirely for several reasons, which include confined placental mosaics,4 co-twin demise,5 maternal chromosomal mosaicism,6 and maternal malignancy.7,8,9
Maternal malignancy, which occurs in approximately 1 per 1000 pregnancies,10 has been associated with the presence of multiple chromosomal aneuploidies in NIPS tests.7,8,11,12,13 One study reported that among 125,426 pregnancies, mothers in 10 pregnancies had maternal malignancies and showed aneuploidy of chromosomes 13, 18, 21, X, or Y in NIPS tests.7 Eight of those mothers showed nonspecific gains and losses of copy number across multiple chromosomes.7 Another study found that among 43 mothers with multiple chromosomal aneuploidies, 18 had maternal malignancy as well as altered genomic profiles.13
These studies suggest that multiple chromosomal aneuploidies may be associated with maternal malignancy. However, it remains difficult for clinicians to comprehend multiple chromosomal aneuploidies findings in NIPS tests and to share the information responsibly with the patient. The objective of this study was to verify the hypothesis that it might be possible to exploit multiple chromosomal aneuploidies findings from NIPS tests and combine them with other cancer markers to detect occult maternal malignancies more reliably. We retrospectively examined test records of more than 1.93 million pregnant women who underwent NIPS tests and identified 639 who tested positive for multiple chromosomal aneuploidies in NIPS tests. We developed a novel cancer detection pipeline (CDP) algorithm that takes into account genome profiles of copy-number variations (CNVs). We also tried to improve the performance of the CDP algorithm with the addition of eight plasma tumor markers.
MATERIALS AND METHODS
Participants
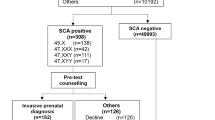
Maternal peripheral blood (10 mL) was collected in Streck Cell Free DNA BCT® blood collection tubes (Streck, La Vista, NE, USA) and processed within four days. NIPS tests and detection of fetal chromosomal aneuploidies were performed as described.14,15,16 Student’s t test was performed based on null/alternative hypotheses, and the relative logarithmic likelihood odds ratio was subsequently calculated. Absolute z-score >3 and L score >1 were used as warning criteria.14,15,16 Multiple chromosomal aneuploidies were defined as two, or more, absolute z-values >3 for any of the 22 autosomes or the X chromosome. We retrospectively reviewed test records of more than 1.93 million pregnant women who underwent NIPS tests between January 2016 and December 2017 at BGI Clinical Laboratories in Shenzhen, Wuhan, and Hong Kong in China. In total we found 639 women who tested positive for multiple chromosomal aneuploidies on an initial NIPS test.
Then, 542 of 639 participants were retrospectively interviewed by physicians by telephone and online questionnaire in July and December 2017, June and November 2018 (Supplementary Methods 1). Clinical information regarding patient medical information was mostly obtained from their clinicians using a questionnaire (Supplementary Methods 2). In rare cases physicians were reluctant to collect that information, so it was directly obtained from participants using the aforementioned questionnaire. To avoid causing anxiety for those participants during pregnancy, the follow-up was performed with extreme caution. Physicians were requested not to mention any explicit cancer-related issues in the telephone interview. Participants were classified as having maternal malignancies based on the confirmed medical record within one-year NIPS tests. Noncancer participants were regarded as the participants with multiple chromosomal aneuploidies results but no cancer identified at the last time of follow-up.
This study was in compliance with strict confidentiality guidelines and regulations regarding personal data protection, and it was approved by the Institutional Review Board of BGI (BGI-IRB 17053). All participants provided written informed consent. BGI was granted permission to anonymously use NIPS sequencing data and plasma of participants for research purposes.
Stratification by multiple chromosomal aneuploidies findings in NIPS tests
Women who tested positive for multiple chromosomal aneuploidies in one NIPS test were routinely advised to undergo a second NIPS test with an independent blood sample as soon as possible. So the majority of participants had two records of NIPS tests. Participants were stratified into those who tested positive for multiple chromosomal aneuploidies in two independent NIPS tests with different blood samples (reproducible multiple chromosomal aneuploidies, 419 women), those who tested positive in the first NIPS but negative in the follow-up NIPS test (non-reproducible multiple chromosomal aneuploidies, 129 women), and those for whom no follow-up NIPS test was performed (uncertain multiple chromosomal aneuploidies, 91 women). For participants with reproducible and uncertain multiple chromosomal aneuploidies results, the results were reported as test failure to women and their physicians. For participants with nonreproducible multiple chromosomal aneuploidies, the results of the second tests were reported.
Development and evaluation of the CDP algorithm
Risk of cancer was estimated in participants with multiple chromosomal aneuploidies using a novel CDP algorithm coded in open-source HMMcopy software.17 In brief, sequencing data in FASTQ format were mapped to human genome 19 (hg19) using Burrows–Wheeler Aligner software (Supplementary Methods 3).18 The HMMcopy software took in a bam file from an individual and predicted CNVs using a hidden Markov model (Supplementary Method 4).17 The bin size of CNV analysis used in this study was 1 M. To detect participants with abnormal profiles of CNVs, the fraction of significant CNVs (FCNV) in a non-neutral state was computed across the entire genome using all CNVs except for those in the Y chromosome in each NIPS test. FCNV = number of significant CNV segments/Total number of segments. FCNV scores for participants with reproducible and nonreproducible multiple chromosomal aneuploidies were obtained by averaging results for the two NIPS tests. The CDP model was built based on the 41 participants with cancer and 501 non-cancer participants. Detailed information regarding the development and validation of the CDP model is presented in the Supplementary Methods 5, 6, and 7.
Analysis of plasma tumor markers (PTMs)
Tumor marker tests have been widely implemented in clinical settings to increase the accuracy of diagnosing several cancer types, as well as to monitor disease progression.19 To find out whether tumor markers may improve the accuracy of diagnosing maternal malignancies and provide possible tumor origins, plasma for 466 participants was retrospectively tested for the presence of eight tumor markers using a microarray enzyme-linked immunoassay (BGI-GBI Biotech, Beijing, China) following the manufacturer’s instructions. The plasma of these participants was left over from NIPS tests and was stored in EP tubes. The following PTMs were assayed (with the respective cutoff value recommended by the assay manufacturer): carbohydrate antigen 15-3 (CA15-3), 28 U/ml; ɑ-fetoprotein (AFP), 500 ng/ml; carcinoembryonic antigen (CEA), 5 ng/ml; carbohydrate antigen 19-9 (CA19-9), 37 U/ml; carbohydrate antigen 125 (CA125), 36 U/ml; cancer antigen 72-4 (CA72-4), 10 U/ml; human cytokeratin fragment antigen 21-1 (CYFRA21-1), 3.3 ng/ml; and squamous cell carcinoma antigen (SCC), 1.2 ng/ml. Participants were considered PTM-positive when the concentration of at least one PTM exceeded the prespecified cutoff value. Notably, the reference ranges for these analytes may vary between plasma and serum.
Statistical analyses
Data are presented as the mean (standard deviation, SD) or median (interquartile range, IR). When appropriate, 95% confidence intervals (95% CI) are also reported. Differences in FCNV scores between cancer and non-cancer groups were assessed for significance using the Wilcoxon rank-sum test in R. Differences in FCNV scores between cancer patients in different stages of disease were assessed using the Kruskal–Wallis test. Potential correlations between FCNV scores and interval from NIPS tests to diagnosis of cancer were explored using Spearman correlation analysis in R. Kaplan–Meier diagnostic curves were plotted, and noncancer rates were compared between participant groups using the log-rank test.20 P < 0.05 was predefined to indicate a statistically significant difference.
RESULTS
Overview of maternal malignant cancer cases identified in 639 participants
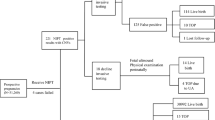
Of the 639 participants enrolled in the study, mean age was 32.1 years (SD, 5.6 years) and mean gestational age was 17 weeks (SD, 3.3 weeks). A total of 542 (84.8%) were interviewed, with a median follow-up of 399.0 days (IR, 302.8–487.0 days) (Fig. 1). While the majority of interviewed participants remained asymptomatic, the median of follow-up was 416.0 days for 501 noncancer participants (IR, 322.0–487.0 days). A total of 41 maternal malignant cancer cases were identified, with 36 from mainland China, 1 from Hong Kong, 2 from Spain, 1 from Taiwan, and 1 from Slovenia. It is noteworthy that 6 of the 41 patients had a cancer diagnosis prior to their NIPS tests (Table S1). Four leiomyomas and one chocolate cyst of ovary were also reported (Fig. 1). At the time of receiving NIPS tests, the 41 cancer patients had a mean age of 33.6 years (SD, 6.3 years) and a mean gestational age of 18.6 weeks (SD, 4.5 weeks). The 41 patients presented a wide spectrum of cancer types, the most frequent being breast cancer (10 cases), liver cancer (9), and lymphoma (9) (Fig. 2a and Table S1). The patients were diagnosed at stage II (8), stage III (9), or stage IV (18), while staging was unknown for the remaining six patients (Fig. 2b, Table S1). Among the 39 patients with cancer for whom the timing of clinical diagnosis was known, the interval from NIPS tests to diagnosis ranged from 0 to 366 days, with a median duration of 115 (IR, 35–192.5 days) (Table S1).
Enrollment of participants, clinical follow-up, identification of maternal malignancy cases, classification of multiple chromosomal aneuploidies results, and development of the CDP model. A total of 639 participants with multiple chromosomal aneuploidies on initial NIPS test from over 1.93 million pregnant women who underwent NIPS tests were enrolled. Analyses of plasma DNA profiles were retrospectively performed using CDP in 639 participants, among whom 466 interviewed participants underwent plasma tumor marker tests. A total of 41 cancer cases, 4 leiomyomas, and 1 case of chocolate cyst of ovary were confirmed. The CDP model was developed with 41 cancer and 501 noncancer participants. CDP cancer detection pipeline built with the software HMMcopy to detect maternal malignancies using NIPS test sequencing data, NIPS noninvasive prenatal screening, PTMs plasma tumor markers.
Reproducible multiple chromosomal aneuploidies are an indicator of cancer risk in NIPS tests
For participants undergoing the second NIPS test, the time interval ranges from 7 to 63 days between the first and second NIPS test, with an average interval of 12 days. Multiple chromosomal aneuploidies findings of any type (reproducible, non-reproducible, or uncertain) were associated with a positive predictive value (PPV) of 7.6% (41/542) for diagnosing maternal malignancy. Of the 41 women with cancer, 34 had reproducible multiple chromosomal aneuploidies and 7 had uncertain multiple chromosomal aneuploidies. Of the 501 noncancer participants, 325 women had reproducible multiple chromosomal aneuploidies, and 176 had non-reproducible or uncertain multiple chromosomal aneuploidies results. Reproducible multiple chromosomal aneuploidies showed diagnostic sensitivity of 82.9% (95% CI, 67.9–92.6%), specificity of 35.1% (95% CI 31.0–39.5%), and PPV of 9.5% (95% CI 8.2–10.9%) (Table S2).
No case of malignancy was found among the 108 participants with non-reproducible multiple chromosomal aneuploidies. These results demonstrate that multiple chromosomal aneuploidies positivity in NIPS tests, particularly reproducible multiple chromosomal aneuploidies, is an indicator of maternal malignancy risk. In contrast, non-reproducible multiple chromosomal aneuploidies may not be associated with maternal malignancies.
Either trisomies or monosomies were shown in both the cancer and noncancer participants. The trisomies occurred most frequently in chromosome 8, 7, and 1 within the maternal malignancy patients, while the most common chromosomal trisomies occurred in chromosome 1, 19, and X within non-cancer participants. The cancer patients showed monosomies in different chromosomes, with chromosome 4, 18, and 13 being most common ones, while the most common chromosomal monosomies occurred in chromosome 19, X, and 16 within noncancer participants (Fig. S1).
CDP is a feasible approach to identify maternal malignancies in multiple chromosomal aneuploidies–positive participants using genome-wide CNV profiles
We examined whether patterns of CNVs differed significantly among pregnant women with multiple chromosomal aneuploidies and malignancies, women with multiple chromosomal aneuploidies but no malignancy, and women without multiple chromosomal aneuploidies. To do this, we compared FCNV scores among 41 cancer patients, 501 non-cancer participants, and 1163 non–multiple chromosomal aneuploidies controls randomly selected from among over 1.93 million pregnant women. The three groups did not differ significantly in age or gestational age (P > 0.05 for all cases, Wilcoxon rank-sum test; Table S3). Cancer patients showed significantly higher FCNV scores than non-cancer participants and non-multiple chromosomal aneuploidies controls (P < 0.0001 for all cases, Wilcoxon sum rank test; Figure 3a, Table S3). Moreover, 39 cancer patients showed abnormal CNVs in NIPS tests (Fig. S2). These results suggest that genomic stability is severely disrupted in maternal malignancies.
The CDP model was built using data from 41 cancer patients and 501 noncancer participants. The optimal cutoff value of the FCNV score was determined to be 0.22. The CDP model allowed prediction of 34 of the 41 (83%) multiple chromosomal aneuploidies women with maternal malignancy. Seventy-nine of 501 (15.8%) of the multiple chromosomal aneuploidies women without malignancy had false positive results. This corresponds to overall sensitivity of 83.0% (95% CI, 67.9 to 92.9%) and PPV of 30.1% (95% CI 25.2% to 35.5%, Table 1) in participants with multiple chromosomal aneuploidies results. Seven cancer patients were predicted by the CDP model to be at low cancer risk, but they were diagnosed with cancer (Table S1); conversely, 79 were predicted to be at high risk, yet they were not diagnosed by the last follow-up. This corresponds to an overall specificity of 84.2% (95% CI 80.7 to 87.3%, Table 1) in participants with multiple chromosomal aneuploidies results. Compared with the diagnostic performance of reproducible multiple chromosomal aneuploidies on their own, the CDP model showed similar sensitivity (P > 0.05, Fisher’s exact test; Table S2) but significantly higher specificity and PPV (both P < 0.0001, Fisher’s exact test; Table S2). Among 429 participants at low risk of cancer, only 7 cancer cases were confirmed, giving a negative predictive value (NPV) of 98.4%. The robustness of the CDP model was assessed by randomly generating ten pairs of training and validation sets at the ratio of 6:4, deriving the corresponding CDP models, and comparing the resulting values of sensitivity, specificity, PPV, NPV, and FCNV cutoff. The model showed mean sensitivity of 87% (SD, 9%) and mean specificity of 81% (SD, 4%) in the whole set of 41 cancer patients and 501 noncancer participants with multiple chromosomal aneuploidies results (Table S4). The receiver operating characteristic (ROC) curves for the training and validation sets in one representative iteration are shown in Fig. 3b. These results suggest that CDP is a feasible approach for detecting maternal malignancies among pregnant women with multiple chromosomal aneuploidies results.
Cancer detection pipeline (CDP) analyses of 41 cancer cases in this study. a Comparison of fraction of significant copy-number variation (FCNV) scores among 41 maternal malignancies cases, 501 non-cancer participants, and 1163 non–multiple chromosomal aneuploidies controls. The red line represents the cutoff of 0.22 to determine CDP positivity. Non–multiple chromosomal aneuploidies controls were 1163 pregnant women without multiple chromosomal aneuploidies results who were randomly selected from 1.93 million pregnant women who took a noninvasive prenatal screening (NIPS) test during the period for study eligibility. b Comparison of receiver operating characteristic curves (ROCs) for FCNV scores in one representative iteration of training and validation sets. c Comparison of FCNV scores in maternal malignancy patients at different cancer stages. d Correlation analysis between FCNV scores and the interval from NIPS tests to diagnosis of cancer in 39 cancer patients. The red line is a linear regression line between FCNV scores and the time from NIPS tests to diagnosis of cancer.
Comparison of FCNV scores among cancer patients in different stages revealed no significant differences (chi-squared = 1.25, P = 0.53, Kruskal–Wallis rank sum test, Figure 3c). However, FCNV scores showed a significant negative correlation with interval from NIPS tests to cancer diagnosis in 39 participants with cancer (correlation coefficient = −0.53, P = 5e-04, Spearman correlation; Fig. 3d). These results suggest that it may be helpful to provide timely preventive clinical intervention to pregnant women who receive high FCNV scores after NIPS tests.
The CDP algorithm can be further improved by incorporating PTM testing
Of the 39 cancer patients with PTM results in our sample, 21 showed elevated levels of at least one of the eight PTMs, and all were predicted by the CDP model to be at high risk of malignancy. The remaining 18 had completely normal PTM results, and 11 were predicted by CDP to be at high risk of malignancy (Table S1 and S5).
Of 542 participants for whom follow-up data were available, 103 were predicted by the CDP model to be at high risk of cancer and 28 had elevated levels of at least one PTM; 21 of these 28 patients were diagnosed with cancer. This corresponds to a PPV as high as 75%. The rate of cancer was significantly higher among participants who were at high cancer risk based on the CDP model and who had elevated levels of at least one PTM than that among participants who were not positive for either of these tests (all P < 0.0001, log-rank test; Fig. 4). These results suggest that a pregnant woman who is predicted by the CDP model to be at high risk and who is PTM-positive may be at particularly high risk of having presymptomatic cancer and should undergo further testing in a timely manner.
The Kaplan–Meier diagnostic plot shows non-cancer rates for five subgroups of participants: those positive for multiple chromosomal aneuploidies, those positive by CDP and PTMs, those positive by CDP but negative by PTMs, those negative by both CDP and PTMs, and those negative by CDP but positive by PTMs. The Kaplan–Meier plots were generated for 466 participants using five analytical methods involving CDP in combination with PTM results and clinical follow-up data. The numbers in brackets are the total number of participants and the number of confirmed cancer cases; the numbers at each time point are the number of noncancer participants. The vertical bars show the interval from NIPS tests to follow-up time for noncancer participants. The turns are the cancer cases in which the interval from NIPS tests to diagnosis was shown on the X axis; asterisk (*) denotes the interval from NIPS tests to follow-up time for two cancer cases without diagnostic date. CDP cancer detection pipeline, NIPS noninvasive prenatal screening, PTMs plasma tumor markers.
PTM testing can provide information on tumor type, in contrast to NIPS tests, which rely on profiles of abnormal CNVs from tumor DNA circulating in the plasma. We found that nine of ten breast cancer patients had elevated levels of at least one of the following PTMs: CA15-3, CA19-9, CA12-5, and CEA. Three patients with liver cancer showed AFP values over 1100 ng/ml, while the other six liver cancer patients were AFP-negative. Of the seven patients with gastrointestinal cancer in whom PTMs were assayed, four showed elevated levels of at least one marker. Of the nine participants with lymphoma for whom PTM results were available, eight were normal for all PTMs tested, while one participant had a marginally elevated level of CA12-5. This suggests that PTMs are poor at detecting lymphoma. The lung cancer case showed elevated CA15-3, CA19-9, CA12-5, and CEA levels (Table S5). Taken together, our results suggest that PTM tests may complement CDP for diagnosing both the presence and likely type of maternal malignancies.
DISCUSSION
Multiple chromosomal aneuploidies are relatively a rare finding in NIPS tests. The incidence rate of multiple chromosomal aneuploidies was 0.03% in this study, which is in line with Bianchi’s study. Trisomy 21, 18, and 13, rare chromosome trisomy, and monosomy have been frequently reported, and miscarriage, placental mosaicism, and uniparental disomy are associated with rare autosomal trisomies.21,22,23,24,25 In this study, 41 cases of cancer, 4 cases of uterine leiomyoma, and 1 case of chocolate cyst of ovary have been confirmed. For the remaining participants, the causes of multiple chromosomal aneuploidies remain unknown. The possible causal factors in the formation of multiple chromosomal aneuploidies including confined placental mosaicism should be explored further in our future studies. Notably, multiple chromosomal aneuploidies findings of any type (reproducible, non-reproducible, or uncertain) were associated with a PPV of 7.6% (41/542) for diagnosing maternal malignancies, which is much lower than 17.9% (7/39) reported by Bianchi’s study. The difference might be attributable to differences in participant populations or to NIPS test algorithm design.
Because maternal malignancy is associated with the presence of multiple chromosomal aneuploidies in NIPS tests,7,8,11,12,13 how to comprehend and convey those multiple chromosomal aneuploidies results during pregnancy is critical and needs be to very cautious. A small group of cancer patients may benefit from being informed of their cancer risk based on the multiple chromosomal aneuploidies results, so they can be diagnosed and treated in a timely manner. However, the high false positive rate of multiple chromosomal aneuploidies results will cause unnecessary stress and invasive follow-up testing in the vast majority of pregnant women.
The results of this study suggest that a CDP model based on genome profiling of CNVs can substantially improve the accuracy of detecting asymptomatic cancer in pregnant women who have shown the presence of multiple chromosomal aneuploidies in NIPS tests, and the performance of the CDP model can be further enhanced by combining it with PTM assays. This clinical pathway may help identify cancer cases from NIPS tests more reliably than multiple chromosomal aneuploidies findings on their own.
Because identifying the primary tumor origin can help guide treatment, we turned to PTM assays, which can be more cost-effective than, for example, whole-body magnetic resonance imaging.8,28 We found that detection of elevated levels of any of eight PTMs was sufficient, together with our CDP model, to achieve an extremely high PPV of 75%. These patients should be examined further in a timely manner because they may be at particularly high risk of occult cancer. In this study, five patients showing CDP-positive and abnormal PTMs results were recommended to undergo medical examinations; they were finally diagnosed with breast cancer (3 cases), liver cancer (1 case), and rectal cancer (1 case) (Table S1). However, it may be helpful to recommend further exams for all patients predicted by our CDP model to be at high cancer risk, given that PTMs on their own show a diagnostic sensitivity of only 53.8% (21/39).
Our CDP method analyzes genome profiles of CNVs. The ability of CDP to integrate FASTQ data from NIPS tests presumably allows it to capture changes in the genomic landscape in a comprehensive manner. This may help explain why our CDP model showed much higher specificity and PPV than reproducible multiple chromosomal aneuploidies on their own. Because the FCNV score can be calculated directly from the NIPS test data with no extra cost, the CDP model may give clinicians more information to evaluate the risk of cancer, thus providing more appropriate genetic counseling and medical workups, and substantially increasing the ability of NIPS tests to detect occult maternal malignancies.
We have noticed that Carlson et al. proposed an algorithm to evaluate women with multiple chromosomal aneuploidies results in a recent publication.26 They emphasized the discussion between clinician and laboratory, and they recommended a stepwise evaluation including history, physical and laboratory evaluation, chest radiograph, and magnetic resonance image. We think this algorithm is suitable for the CDP-positive population, which needs extended evaluation, especially with positive PTM results at the same time. Our results suggest the need to carefully consider whether to reveal the possibility of maternal malignancies to pregnant women with negative CDP results, because the PPV of cancer in this group was relatively low 1.6% (7/429). But this algorithm may be still suitable if the clinician thinks the evaluation is necessary. A noninvasive postnatal test to assess genomic profiles without the interference of fetus and placenta (Fig. S3) may be useful, but this possibility should be examined in further studies.
The present work makes evidence-based recommendations for how clinicians can proceed if a NIPS test detects multiple chromosomal aneuploidies in a pregnant woman. Nevertheless, our conclusions should be interpreted with caution in light of several limitations. First, this is a retrospective study and many participants are still under follow-up; the results may change as more cases of cancer are diagnosed. Second, our cancer prevalence of 41 per 1.93 million is significantly lower than a previous report,10 which may reflect the fact that we focused on cancer patients who were positive for multiple chromosomal aneuploidies on an initial NIPS test, rather than on pregnant women with any type of malignancy. This large difference in prevalence suggests that the CDP algorithm may work well for identifying occult maternal malignancies associated with multiple chromosomal aneuploidies, but it may not work as a screening test for all pregnancy-related tumors. Third, multiple chromosomal aneuploidies were derived from the z-score algorithm. The findings and conclusions may be highly specific to the particular NIPS assay system at BGI. Further work should seek to verify and extend our findings in a larger sample of women with confirmed maternal malignancies.27
Conclusion
Our results suggest that reproducible multiple chromosomal aneuploidies detectable in NIPS tests are associated with maternal malignant cancer risk, and that the combination of multiple chromosomal aneuploidies findings with our novel CDP algorithm, based on low-coverage NIPS test sequencing data, shows potential for detecting presymptomatic maternal malignancies. The positive predictive value of the CDP model may be substantially improved by incorporating results from plasma tumor markers, which can also provide information about primary tumor origin. Our findings lay the foundation for applying the NIPS test not just to screen for fetal aneuploidies but also to uncover occult maternal malignancies.
References
Agarwal A, Sayres LC, Cho MK, et al. Commercial landscape of noninvasive prenatal testing in the United States. Prenat Diagn. 2013;33:521–531.
Benn P, Cuckle H, Pergament E. Non-invasive prenatal testing for aneuploidy: Current status and future prospects. Ultrasound Obstet Gynecol. 2013;42:15–33.
Chen EZ, Chiu RWK, Sun H, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma dna sequencing. PLoS ONE. 2011;6:1–7.
Lau TK, Jiang FM, Stevenson RJ, et al. Secondary findings from non-invasive prenatal testing for common fetal aneuploidies by whole genome sequencing as a clinical service. Prenat Diagn. 2013;33:602–608.
Curnow KJ, Wilkins-Haug L, Ryan A, et al. Detection of triploid, molar, and vanishing twin pregnancies by a single-nucleotide polymorphism-based noninvasive prenatal test. Am J Obstet Gynecol. 2015;212:79e1–79e9.
Bianchi DW, Parsa S, Bhatt S, et al. Fetal sex chromosome testing by maternal plasma DNA sequencing: clinical laboratory experience and biology. Obstet Gynecol. 2015;125:375–382.
Bianchi DW, Chudova D, Sehnert AJ, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314:162–169.
Amant F, Verheecke M, Wlodarska I, et al. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol. 2015;1:814–819.
Hartwig TS, Ambye L, Sørensen S, Jørgensen FS. Discordant non-invasive prenatal testing (NIPT)—a systematic review. Prenat Diagn. 2017;37:527–539.
Pavlidis NA. Coexistence of pregnancy and malignancy. Oncologist. 2002;7:279–287.
Sun K, Jiang P, Chan KCA, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci USA. 2015;112:E5503–E5512.
Osborne CM, Hardisty E, Devers P, et al. Discordant noninvasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenat Diagn. 2013;33:609–611.
Dharajiya NG, Grosu DS, Farkas DH, et al. Incidental detection of maternal neoplasia in noninvasive prenatal testing. Clin Chem. 2018;64:329–335. https://doi.org/10.1373/clinchem.2017.277517. [Epub 2017 Oct 5].
Dan S, Wang W, Ren J, et al. Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11105 pregnancies with mixed risk factors. Prenat Diagn. 2012;32:1225–1232.
Lau TK, Chan MK, Lo PSS, et al. Clinical utility of noninvasive fetal trisomy (NIFTY) test—early experience. J Matern Fetal Neonatal Med. 2012;25:1856–1859.
Chen S. et al. A method for noninvasive detection of fetal large deletions/duplications by low coverage massively parallel sequencing. Prenat Diagn.2013;33:584–590.
Ha G, Roth A, Lai D, et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 2012;22:1995–2007.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760.
Borrebaeck CAK. Precision diagnostics: moving towards protein biomarker signatures of clinical utility in cancer. Nat Rev Cancer. 2017;17:199–204.
Terry M. Therneau and Patricia M. Grambsch (2000). Modeling Survival Data: Extending the Cox Model. Springer, New York.
Tørring N, Petersen OB, Becher N, et al. First trimester screening for other trisomies than trisomy 21, 18, and 13. Prenat Diagn. 2015;35:612–619.
Pescia G, Guex N, Iseli C, et al. Cell-free DNA testing of an extended range of chromosomal anomalies: clinical experience with 6,388 consecutive cases. Genet Med. 2017;19:169–175.
Pertile MD, Halks-Miller M, Flowers N, et al. Rare autosomal trisomies, revealed by maternal plasma DNA sequencing, suggest increased risk of feto-placental disease. Sci Transl Med. 2017;9:1–12.
Liang D, Lin Y, Qiao F, et al. Perinatal outcomes following cell-free DNA screening in >32 000 women: Clinical follow-up data from a single tertiary center. Prenat Diagn. 2018;38:755–764.
Dharajiya NG, Namba A, Horiuchi I, et al. Uterine leiomyoma confounding a noninvasive prenatal test result. Prenat Diagn. 2015;35:990–993.
Carlson LM, Hardisty E, Coombs CC, Vora NL. Maternal malignancy evaluation after discordant cell-free DNA results. Obstet Gynecol. 2018;131:464–468.
Amant F, Vandenbroucke T, Verheecke M, et al. Pediatric outcome after maternal cancer diagnosed during pregnancy. N Engl J Med. 2015;373:1824–1834.
Peccatori FA, Codacci-Pisanelli G, Del Grande M, et al. Whole body MRI for systemic staging of breast cancer in pregnant women. Breast. 2017;35:177–181.
Acknowledgements
The study was supported by the National Key Research & Development Program of China (grant number 2016YFC0905400); Young Scientists Fund of the National Natural Science Foundation of China (grant number 81601293); Natural Science Foundation of Guangdong Province, China (2017A030306026); the Fund for Distinguished Young Scholars of South China University of China (2017JQ017); the Guangzhou Science and Technology Project (201400000004-5, 201508020247); the Pearl River Nova Program of Guangzhou (201506010065); National Key R&D Program of China (grant number 2018YFC1002900, 2018YFC1002901); and High-level Talents of Guangdong (grant number 2016TX03R171).
Author information
Authors and Affiliations
Contributions
XJ, JL, YHH, PLS, and YYY are joint first authors. XJ, YW C and MM are joint corresponding authors on this study. All authors were involved in designing the study; following up with patients; collecting, analyzing, and interpreting the data; and writing the manuscript. All genomic data for all participants and computer codes from this study are deposited at China Nucleotide Sequence Archive (accession ID CNP0000067, https://db.cngb.org/cnsa/project/CNP0000067/public/).
Corresponding authors
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ji, X., Li, J., Huang, Y. et al. Identifying occult maternal malignancies from 1.93 million pregnant women undergoing noninvasive prenatal screening tests. Genet Med 21, 2293–2302 (2019). https://doi.org/10.1038/s41436-019-0510-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-019-0510-5
Keywords
This article is cited by
-
Noninvasive prenatal screening and maternal malignancy: role of imaging
Abdominal Radiology (2023)
-
Detection of cryptogenic malignancies from metagenomic whole genome sequencing of body fluids
Genome Medicine (2021)