Abstract
Purpose
Some 10% of familial adenomatous polyposis (FAP) and 80% of attenuated polyposis (AFAP) cases remain molecularly unexplained. We scrutinized such cases by exome-wide and targeted methods to search for novel susceptibility genes.
Methods
Exome sequencing was conducted on 40 unexplained (mainly sporadic) cases with FAP or AFAP from Finland. The DNA mismatch repair (MMR) gene MLH3 (MutL Homolog 3) was pinpointed and prompted a subsequent screen of ~1000 Swedish patients referred to clinical panel sequencing for colon tumor susceptibility.
Results
Three homozygous carriers of a truncating variant in MLH3, c.3563C>G, p.Ser1188Ter, were identified among the index cases from the Finnish series. An additional biallelic carrier of the same variant was present in the Swedish series. All four patients shared a 0.8-Mb core haplotype around MLH3, suggesting a founder variant. Colorectal polyps from variant carriers showed no instability at mono-, di-, tri-, or tetranucleotide repeats, in agreement with previous findings of a minor role of MLH3 in MMR. Multiple loci were affected by loss of heterozygosity, suggesting chromosomal instability.
Conclusion
Our results show that a biallelic nonsense variant of MLH3 underlies a novel syndrome with susceptibility to classical or attenuated adenomatous polyposis and possibly extracolonic tumors, including breast cancer.
Similar content being viewed by others
INTRODUCTION
Familial adenomatous polyposis (FAP, MIM 175100) is characterized by multiple polyps in the colon and rectum and an increased risk of colorectal cancer. Most patients with classical FAP (polyp count at least 100) show heterozygous pathogenic germline variants in the APC gene.1 Alterations in the extreme 5’ or 3’ ends or the alternatively spliced exon 9 of APC are associated with attenuated disease (AFAP, 10–100 polyps). (A)FAP-like polyposis may also result from pathogenic sequence changes in various other genes, including heterozygous variants in POLE and POLD1 (polymerase proofreading-associated polyposis; MIM 615083 and 612591, respectively), or homozygous variants in MUTYH (MIM 608456), NTHL1 (MIM 616415), and MSH3 (MIM 617100) (ref. 1). Significant proportions of polyposis cases (10–80% depending on polyp count) remain molecularly unexplained,2 encouraging searches for novel susceptibility genes.
Accurate molecular and clinical classification of conditions with colorectal polyposis is a prerequisite for appropriate genetic counseling and testing to guide patient management and colorectal cancer prevention. To this end, we undertook an exome sequencing study of 40 index cases from a cohort with adenomatous polyposis from Finland in which pathogenic variants in known polyposis-associated genes had been excluded.2 Results from this cohort, supplemented with panel sequencing data from some 1000 cases with various colorectal phenotypes from Sweden, unravel a novel polyposis syndrome associated with biallelic germline nonsense variant of MLH3 (MutL Homolog 3).
MATERIALS AND METHODS
Patient cohorts and strategies to screen for pathogenic germline variants
The patient series investigated are described in Supplementary Materials and Methods and schematically depicted in Fig. 1a. In the first phase, blood DNAs from 40 unrelated cases/families with adenomatous polyposis from Finland, without any detectable pathogenic sequence changes in known polyposis-associated genes2 and in most cases sporadic at presentation, were submitted to exome sequencing (ES). Based on polyp count (with 100 polyps as a divider), 14 represented FAP and 26 AFAP. Guided by the findings from the Finnish polyposis cohort, 829 patients from Sweden, referred to Cancer Genetics Counseling clinics for a suspected hereditary colorectal cancer syndrome, were analyzed for MLH3 variants from panel sequencing data. Variants with allele frequency below 0.003 (gnomAD), nonsynonymous (frameshift, stop gained/lost, missense, disrupting donor/acceptor site variants), and predicted pathogenic with at least five of six programs assessing protein function in silico were selected. The recurrent MLH3 c.3563C>G, p.Ser1188Ter variant was further characterized by studying constitutional DNA (haplotype analyses for shared versus independent origin) and RNA (primer extension and cloning analyses for nonsense-mediated RNA decay). Additionally, tumor tissues from variant carriers were examined for microsatellite instability (MSI), loss of heterozygosity (LOH)/allelic imbalance (AI), CpG island methylator phenotype (CIMP), and MMR protein expression (see Supplementary Materials and Methods). Written informed consent preceded study participation and sample donation. This study was approved by the institutional review board of the Helsinki University Central Hospital (Helsinki, Finland) and by the local ethics committees of the Universities of Gothenburg and Lund, Sweden.
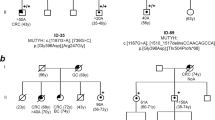
(a) Flow-chart of this investigation including the Finnish and Swedish arms. MLH3 findings and methods used for their identification are shown. (b) Pedigrees of polyposis families with the MLH3 p.Ser1188Ter variant. Families 158, 168, 177, and 1007 are from Finland and family SWE is from Sweden. The pedigrees were generated with Pedigree Chart Designer. Numbers below the symbols are patient identifiers. Arrow denotes the index person. Carrier status for the MLH3 c.3563C>G, p.Ser1188Ter variant is shown (+/+, homozygous carrier, +/− heterozygous carrier). Tumor manifestations and age at diagnosis (years) are given below the patient symbol. AFAP attenuated familial adenomatous polyposis, ES exome sequencing, FAP familial adenomatous polyposis, LS Lynch syndrome.
RESULTS
Identification of a recurrent germline variant in MLH3
Forty index cases with unexplained adenomatous polyposis from Finland were investigated by exome sequencing to dissect the underlying genes (Fig. 1a). After filtering out variants common in the population and those unlikely to be pathogenic (see “Materials and methods”), a recurring nonsense variant affecting the DNA mismatch repair (MMR) gene MLH3 (c.3563C>G, p.Ser1188Ter) (Table S1) was observed in four families (Fig. 1b). The index individuals from families 158, 168, and 177 were homozygous for the variant (Figure S1). Polyposis was attenuated in the first one and profuse in the latter two. The index person from family 1007 was heterozygous for the variant and showed mild polyposis. The variant is absent in disease- or locus-specific databases and rare in the population (allele frequency 0.0002603 in gnomAD and 0.00238 in Finns based on SISu; no homozygotes reported).
Our subsequent literature search identified a recent study on 91 Swedish cases referred to clinical panel sequencing for FAP or Lynch syndrome (LS, MIM 120435 and related).3 A sporadic case with apparently classical FAP (case IV:69 [ref. 3], who is the index individual of SWE family in Fig. 1b) was diagnosed with the homozygous MLH3 p.Ser1188Ter variant. Prompted by this finding, the entire MLH3 gene was investigated in an additional cohort of 829 Swedish cases referred to clinical laboratory screening for a suspected hereditary colorectal cancer syndrome (Fig. 1a). No additional cases with homozygous pathogenic or likely pathogenic MLH3 variants were found, and no concomitant nonsynonymous variants in MLH3 were detected in any of the patients (Table S2). Despite looking like unrelated families, the four Finnish cases and the single Swedish case with the MLH3 p.Ser1188Ter variant all shared a 0.8-Mb haplotype around MLH3 (Fig. 2a), suggesting origin from a common ancestor. The variant was absent in cohorts of nonpolypotic colon cancer (Fig. 1a).
(a) Shared founder haplotype in carriers of the MLH3 p.Ser1188Ter variant. The haplotypes for an 8.8-Mb region between microsatellite markers D14S268 and D14S1000 are shown for the index patients from the indicated families (158, 168, 177, and SWE, homozygous carriers; 1007, heterozygous carrier). The conserved disease haplotype associated with the p.Ser1188Ter variant (between positions 73,271,847 and 75,777,880) is shaded. Deviation from the conserved haplotype at D14S1047 in SWE could result from either mutation or recombination. A core haplotype of 0.8 Mb (between D14S1047 and D14S284) that includes the MLH3 gene and is shared by all p.Ser1188Ter variant carriers is indicated by brackets. (b) Location of the p.Ser1188Ter variant relative to the main functional domains of MLH3. A lollipop diagram of the MLH3 protein was created by MutationMapper. The functional domains are as follows: ATPase domain, MMR (DNA mismatch repair domain), and MutL_C (MutL C terminal dimerization domain). (c) Results from microsatellite instability (MSI) and allelic imbalance (AI)/loss of heterozygosity (LOH) analyses of colorectal adenomas from MLH3 p.Ser1188Ter variant carriers. The markers used were BAT25 (mononucleotide repeat from chromosome 4), D17S250 (dinucleotide repeat from chromosome 17), and D9S242 (tetranucleotide repeat from chromosome 9). All tumors were microsatellite-stable (MSS, marker BAT25). Allelic imbalance was evident with D17S250 and D9S242.
Molecular characteristics of the MLH3 founder variant
The MLH3 p.Ser1188Ter variant causes an immediate stop of translation, leading to the loss of the MLH1 binding domain of MLH3 (Fig. 2b). The variant was associated with nonsense-mediated RNA decay by primer extension and cloning experiments (Figure S2). While none of the commercially available MLH3 antibodies tested for immunohistochemistry produced staining patterns of sufficient specificity, MSI results were unequivocal: no single tumor (adenoma or carcinoma, among a total of eight tumors investigated) from the MLH3 p.Ser1188Ter variant carriers revealed any MSI at mono-, di-, tri-, or tetranucleotide repeats, either by conventional polymerase chain reaction (PCR) (Fig. 2c) or small pool PCR. Instead, 8 of 15 di-, tri-, and tetranucleotide markers tested showed loss of heterozygosity or allelic imbalance in colorectal and other tumors (including breast carcinoma), suggesting chromosomal instability (Fig. 2c). Immunohistochemical analysis showed normal expression of MLH1 protein, the binding partner of MLH3 (ref. 4). One adenoma of three tested revealed a BRAF-V600E variant in association with CpG island methylator phenotype (see Supplementary Materials and Methods).
Concurrent sequence changes in other genes from MLH3 variant carriers
Apart from the biallelic p.Ser1188Ter variant in MLH3, the index patient from family 158 showed two MSH3 variants, a truncating (c.1308_1309delAG, p.Glu437fs) and a missense change (c.2692A>C, p.Asn898His) (Table S1). However, a complementary DNA (cDNA) cloning experiment showed that both variants affected the same allele; moreover, MSH3 protein was present in tumor tissue by immunohistochemical analysis (Figure S3). High-penetrance heterozygous pathogenic germline variants of MSH3 are very rare in individuals with a suspected cancer predisposition.5 Furthermore, family 158 with microsatellite-stable tumors showed no evidence of a possible synergistic effect with other MMR gene variants, as proposed to occur in some Lynch families.6 Therefore, heterozygosity for the MSH3 variants in family 158 is likely to represent a secondary finding. The index case from family 168 carried a potentially pathogenic7 splice variant in CHEK2 (c.319+2T>A), but it was absent in her affected siblings. The lack of additional convincingly or likely pathogenic germline variants by ES suggested that the biallelic MLH3 p.Ser1188Ter variant was the primary alteration behind polyposis in families 158, 168, and 177.
MMR gene variants found in index cases without the MLH3 founder variant
Based on clinical presentation as polyposis patients, previous diagnostic tests conducted on the 40 polyposis cases from Finland focused on established polyposis-associated genes. Our exome sequencing experiments identified rare heterozygous variants in the LS predisposition gene MSH2 in two cases. Both variants involved the evolutionarily conserved MSH3/MSH6 interaction domain of the MSH2 gene product and were absent in disease- or locus-specific databases. The index individual from family 1001 (with polyp count 10 and a personal and family history of colorectal cancer) had a 27-bp in-frame deletion (c.1140_1166del, p.Leu381_Arg389del) likely to have pathogenic significance because it was accompanied by MSI-high and absent MSH2 and MSH6 proteins in tumor tissue. The index individual from family 1025 (a sporadic case with polyp count 20–30) revealed a missense change (c.1337A>G, p.Asp446Gly) predicted deleterious by all six in silico programs tested. However, as the variant was associated with stable microsatellites and normal MMR protein expression in tumor tissue, its significance remains unknown.
DISCUSSION
We describe a novel polyposis and cancer syndrome associated with biallelic (homozygous) pathogenic germline variant of the “minor” MMR gene MLH3. MLH3 was originally identified as a colon tumor susceptibility gene in mice some 20 years ago.8 A few dozen heterozygous germline variants in MLH3 with possible pathogenicity, mostly missense but also truncating, are listed in the InSiGHT database (www.insight-group.org). The clinical significance of such variants remains unsettled; they may play a role in familial nonpolypotic colon cancer or LS predisposition (MIM 614385) (refs. 9,10). MSI findings from tumor tissues vary from instability at primarily di- and tetranucleotide repeats9 to no MSI.10 Cosegregation results are variable and a role as a low-penetrance colon cancer susceptibility gene has been suggested.10,11 Due to the lack of genome-wide sequencing data, concurrent pathogenic variants in other cancer-relevant genes cannot often be excluded. No increased risk of colorectal adenomas in putative LS families with MLH3 variants has been reported.9,10
Our results link a biallelic germline nonsense variant of MLH3 to a clinical and molecular phenotype different from LS: microsatellite-stable adenomatous polyposis exhibiting chromosomal instability. Whether or not the rate of LOH/AI in MLH3 variant carriers exceeds that observed in the corresponding sporadic tumors remains to be addressed by future studies. Moreover, the somatic BRAF-V600E variant was detected in one of three adenomas from MLH3 p.Ser1188Ter variant carriers, which is another discriminating feature since BRAF-V600E generally predicts the absence of LS.12 The lack of MSI in the MLH3 variant carriers is compatible with the reported minor role for the MLH1-MLH3 complex (hMutLγ) in MMR, reflecting redundancy with the main MMR complex MLH1-PMS2 (hMutLα).4 The accumulated data from yeast, mouse, and human cells suggest a primary function for MLH3 in meiotic recombination instead of MMR.13,14,15 Extrapolation from literature data allows us to hypothesize that relevant functions that may fail in the MLH3 p.Ser1188Ter-associated tumorigenesis might involve, for example, DNA damage response (defective in Mlh3−/− mice)14 or recombination-related processes.16
LS associated with “major” MMR genes MLH1, MSH2, MSH6, and PMS2 can occasionally present with polyposis. Two index cases from the Finnish polyposis series (2/40, 5%), both with attenuated polyposis, revealed novel MMR gene variants in MSH2 by exome sequencing. The findings comply with Kalady et al.17 and suggest that although ten or more adenomas prompt testing for polyposis, the possibility of LS should not be overlooked.
While heterozygous sequence changes of MLH1, MSH2, MSH6, and PMS2 can cause predisposition to LS, biallelic inactivation of the same genes underlies constitutional MMR deficiency syndrome (CMMRD; MIM 276300). CMMRD is characterized by variable penetrance and diverse clinical manifestations, including colonic adenomatous polyposis of variable degree.18 Clinically, the MLH3-associated syndrome we describe shares certain CMMRD features in analogy to the MSH3-associated polyposis syndrome identified recently.5 The age at onset of polyposis (classical or attenuated) was relatively late (48–52 years in the biallelic index cases), which might suggest reduced penetrance. The cases were considered sporadic relative to polyposis, but the family histories did include various noncolonic cancers, especially breast cancer (Fig. 1b). Breast cancer co-occurred with polyposis in the index patient (III.3) from family 168 and was the only tumor manifestation in her sister (III.2), likewise homozygous for the MLH3 founder variant. While the possibility of the (postmenopausal) breast carcinomas being phenocopies cannot be excluded, heterozygous (truncating) germline variants in MLH3 identified by recent massive parallel sequencing studies of breast cancer patients support an association between MLH3 and breast cancer.19 The observation of brain tumors in the mothers of the index cases of families 158 and 168 is also of potential interest given that multiple MLH3 variants have been reported to coexist in colon cancer patients from families with LS-associated brain tumors.20 The fact that none of the parents of our homozygous polyposis patients were affected with polyposis is consistent with recessive inheritance; however, two proven heterozygous individuals had a disease phenotype (attenuated polyposis in II.2 from family 1007 and renal urothelial cancer in III.1 from family 168). Because our data rely on a founder variant enriched in the Finnish population, investigations on additional cohorts and populations are warranted to establish the significance of MLH3 as a polyposis-predisposing factor and to define the tumor spectrum of the syndrome.
References
Basso G, Bianchi P, Malesci A, Laghi L. Hereditary or sporadic polyposis syndromes. Best Pract Res Clin Gastroenterol. 2017;31:409–417.
Nieminen TT, Pavicic W, Porkka N, et al. Pseudoexons provide a mechanism for allele-specific expression of APC in familial adenomatous polyposis. Oncotarget. 2016;43:70685–70698.
Rohlin A, Rambech E, Kvist A, et al. Expanding the genotype-phenotype spectrum in hereditary colorectal cancer by gene panel testing. Fam Cancer. 2017;16:195–203.
Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5:a012633.
Adam R, Spier I, Xhao B, et al. Exome sequencing identifies biallelic MSH3 germline mutations as a recessive subtype of colorectal adenomatous polyposis. Am J Hum Genet. 2016;99:337–351.
Duraturo F, Liccardo R, Cavallo A, et al. Association of low-risk MSH3 and MSH2 variant alleles with Lynch syndrome: probability of synergistic effects. Int J Cancer. 2011;129:1643–1650.
Dominguez-Valentin M, Nakken S, Tubeuf H, et al. Potentially pathogenic germline CHEK2 c.319+2T>A among multiple early-onset cancer families. Fam Cancer. 2018;17:141–153.
Lipkin SM, Wang V, Jacoby R, et al. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet. 2000;24:27–35.
Wu Y, Berends MJ, Sijmons RH, et al. A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nat Genet. 2001;29:137–138.
Liu HX, Zhou XL, Liu T, Werelius B, Lindmark G, Dahl N, Lindblom A. The role of hMLH3 in familial colorectal cancer. Cancer Res. 2003;63:1894–1899.
Korhonen MK, Vuorenmaa E, Nyström M. The first functional study of MLH3 mutations found in cancer patients. Genes Chromosomes Cancer. 2008;47:803–809.
Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49:151–157.
Bouuaert CC, Keeney S. Distinct DNA-binding surfaces in the ATPase and linker domains of MutLγ determine its substrate specificities and exert separable functions in meiotic recombination and mismatch repair. PLoS Genet. 2017;13:e1006974.
Chen PC, Dudley S, Hagen W, et al. Contributions by MutL homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65:8662–8670.
Cannavo E, Marra G, Sabates-Bellver J, et al. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–10766.
Jahid S, Sun J, Gelincik O, et al. Inhibition of colorectal cancer genomic copy number alterations and chromosomal fragile site tumor suppressor FHIT and WWOX deletions by DNA mismatch repair. Oncotarget. 2017;8:71754–71586.
Kalady MF, Kravochuck SE, Heald B, Burke CA, Church JM. Defining the adenoma burden in lynch syndrome. Dis Colon Rectum. 2015;58:388–392.
Durno C, Boland CR, Cohen S, et al. Recommendations on surveillance and management of biallelic mismatch repair deficiency (BMMRD) syndrome: a consensus statement by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2017;152:1605–1614.
Castéra L, Krieger S, Rousselin A, et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet. 2014;22:1305–1313.
Duraturo F, Liccardo R, Izzo P. Coexistence of MLH3 germline variants in colon cancer patients belonging to families with Lynch syndrome-associated brain tumors. J Neurooncol. 2016;129:577–578.
Acknowledgements
We thank the patients and clinical staff for participation. Saila Saarinen is thanked for expert technical assistance and Beatriz Alcala-Repo for collecting clinical information. This work was supported by grants from the Jane and Aatos Erkko Foundation (to T.T.N. and P.P.), the Academy of Finland (grant number 294643, to P.P.), the Finnish Cancer Organizations (to P.P.), the Sigrid Juselius Foundation (to P.P.), the HiLIFE Fellows 2017–2020 (to P.P.), Maud Kuistila Foundation (to T.T.N.), and Swedish Cancer Society (to M.N.) (grant number CAN 2016/645; The study was financed by grants from the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement (ALFGBG-725011).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
SWEN investigators contributing to this study: Gustav Silander, Ekaterina Kuchinskaya, Christos Aravidis, Theofanis Zagoras, Mef Nilbert, and Åke Borg.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. If you remix, transform, or build upon this article or a part thereof, you must distribute your contributions under the same license as the original. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/.
About this article
Cite this article
Olkinuora, A., Nieminen, T.T., Mårtensson, E. et al. Biallelic germline nonsense variant of MLH3 underlies polyposis predisposition. Genet Med 21, 1868–1873 (2019). https://doi.org/10.1038/s41436-018-0405-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0405-x
Keywords
This article is cited by
-
The complementary roles of genome-wide approaches in identifying genes linked to an inherited risk of colorectal cancer
Hereditary Cancer in Clinical Practice (2023)
-
Genotype–Phenotype Correlations in Autosomal Dominant and Recessive APC Mutation-Negative Colorectal Adenomatous Polyposis
Digestive Diseases and Sciences (2023)
-
Use of sanger and next-generation sequencing to screen for mosaic and intronic APC variants in unexplained colorectal polyposis patients
Familial Cancer (2022)
-
Danish guidelines for management of non-APC-associated hereditary polyposis syndromes
Hereditary Cancer in Clinical Practice (2021)
-
A loss-of-function variant in DNA mismatch repair gene MLH3 underlies severe oligozoospermia
Journal of Human Genetics (2021)