Abstract
Purpose
To identify the economic impact of pediatric patients with clinical indications of genetic disease (GD) on the US health-care system.
Methods
Using the 2012 Kids’ Inpatient Database, we identified pediatric inpatient discharges with International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) codes linked to genetic disease, including well-established genetic disorders, neurological diseases, birth defects, and other physiological or functional abnormalities with a genetic basis. Cohort characteristics and health-care utilization measures were analyzed. Discharges with a GD-associated primary diagnosis were used to estimate the minimum burden; discharges with GD-associated primary or secondary codes established the maximum burden.
Results
Of 5.85 million weighted discharges, 2.6–14% included GD-associated ICD-9-CM codes. For these discharges, mean total costs were $16,000–77,000 higher (P < 0.0001) in neonates and $12,000–17,000 higher (P < 0.0001) in pediatric patients compared with background, corresponding to significantly higher total charges and lengths of stay. Aggregate total charges for suspected GD accounted for $14 to $57 billion (11–46%) of the “national bill” for pediatric patients in 2012.
Conclusion
Pediatric inpatients with diagnostic codes linked to genetic disease have a significant and disproportionate impact on resources and costs in the US health-care system.
Similar content being viewed by others
INTRODUCTION
Standard diagnostic approaches to suspected genetic disease (GD) include examination for distinctive clinical finding patterns, followed by narrowly selected molecular tests, to arrive at a precision diagnosis. For example, microarrays have traditionally been utilized for patients with birth defects and neurological disorders,1,2,3 and single-gene testing is often implemented for high penetrance Mendelian conditions. Both approaches are limited, however, when a patient presents with ambiguous signs and symptoms, and may have one of approximately 5000 conditions for which a disease gene has been identified.4 This can lead to long and costly diagnostic odysseys in which these patients remain undiagnosed for several years.
The overall health-care utilization of patients with a possible GD in the United States has not yet been comprehensively assessed, limiting our understanding of the potential impact of new technologies, including genome and exome sequencing,5,6,7,8 as aids to earlier diagnosis. Several barriers, including clinical heterogeneity, incomplete utilization of genetic testing, inconsistent coding practices, and general lack of physician awareness or expertise, make it difficult to capture the incidence and burden of pediatric genetic disease. As a surrogate to identify a study population of patients likely to include most patients with GD, we have used the presence of clinical diagnoses that are typical indications for use for standard genetic tests. These descriptive diagnoses include morphological, physiological, and functional abnormalities that while themselves are not pathognomonic for a genetic diagnosis, are sufficiently uncommon or specific to constitute a reasonable indication for genetic testing9,10,11,12 (also see Table S1). Investigation of these patients’ health-care utilization shows that they require disproportionate resources for their care and highlights the potential for earlier molecular diagnosis to relieve some of the economic burden through reduction in iterative diagnostic approaches and improved targeted interventions.
MATERIALS AND METHODS
Data source
We utilized the 2012 version of the Kids’ Inpatient Database (KID), developed by the Healthcare Cost and Utilization Project (HCUP), sponsored by the Agency for Healthcare Research and Quality (AHRQ). KID is a nationally representative sample and the largest comprehensive, all-payer database for children in the United States.13 KID represents a stratified sampling of HCUP’s State Inpatient Databases, with random sampling of 10% of uncomplicated (healthy) live births and 80% of complicated in-hospital births and other pediatric discharges (age 0 to 20 years).14 Similar to other HCUP national databases, KID data was weighted to produce national and regional estimates.15 Weighted estimates of discharge counts are shown in all result tables and figures. KID is released triennially, and the 2012 version was sampled from over 4000 US community, nonrehabilitation hospitals with pediatric discharges in 46 HCUP member states.16 Patients less than 18 years of age at time of admission in 2012, represented by over 5 million weighted inpatient discharges, were selected for analysis. Diagnoses and procedures were coded using the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM). Research is exempt from Human Subjects Research under OHRP 45 CFR 46.101(b)(2) from all 45 CFR part 46 requirements.
Study population
We aimed to identify discharges in which the clinical diagnoses are acceptable indications for use of genetic testing. An expert medical geneticist compiled a list of 919 ICD-9-CM codes (October 2011), which enabled identification of codes definitely indicative of a genetic disorder (e.g., cystic fibrosis or specific inborn errors of metabolism), likely to be caused by a genetic disorder (i.e., a genetic diagnosis is likely to be achieved with appropriate genetic testing in >50% of discharges with these codes; e.g., neonatal intractable epilepsy), or possibly associated with a genetic disorder (i.e., >10% of discharges will yield a genetic diagnosis with appropriate testing, including most severe birth defects, such as congenital central nervous system [CNS] and cardiac malformations). These ICD-9-CM diagnostic codes (Table S2) represent morphological, physiological, or functional diagnoses that could be used to support a medical decision to undertake genetic testing, and represent a patient population that encompasses those with rare, undiagnosed, or genetic disease.
Table S3 shows the results of a primary literature search supporting the compilation of these codes. Each ICD-9-CM diagnostic code description in Table S2 was parsed to remove nonalphanumeric characters and queried against the Human Phenotype Ontology (HPO)17 and the OMIM4 GeneMap application programming interface (API). OMIM GeneMap and HPO query results were parsed to identify disease names and gene/locus symbols. The resulting ICD-9-CM description, gene/locus symbols, and disease name were queried against a Google custom search engine (CSE) API focused to academic literature using PubMed-listed uniform resource locators (URLs). Google CSE query results were parsed to identify academic publication titles, URLs, and PubMed IDs (PMIDs) that matched the specified query. A medical geneticist manually reviewed the results. While not comprehensive, the literature review (Table S3) shows that the codes in Table S2 can reasonably serve as correlates of genetic disorders for the purpose of this analysis and that their presence is positively associated with genetic diagnoses. Codes were also categorized into 13 ICD-9-CM disease categories for aggregation and analysis, as shown in Table S2.
The maximum number of diagnoses and procedures recorded for each inpatient discharge varies by HCUP member state, but up to 15 diagnoses and 15 procedures were included per discharge in the KID data set and coded in ICD-9-CM. In the HCUP data, the first diagnosis is considered the primary diagnosis, and all other ICD-9-CM codes are considered secondary. Using the 919 ICD-9-CM codes, we identified two sets of criteria by which to define the prevalence range. To establish the upper bound of possible GD discharges, we defined the maximum number of discharges of patients with suspected GD as those for which any diagnosis (up to 15 diagnoses per discharge) was included in the list of 919 codes. For a conservative minimum, discharges with a primary diagnosis in the set of ICD-9-CM codes were selected. In HCUP and other claims data sets, the first listed diagnosis is commonly defined as the condition to be chiefly responsible for inpatient admission and care.18,19 To account for variability in coding practice and discharges for newborns, in which “live birth” is often coded first, we considered any diagnosis in our maximum definition, as has been done in similar studies.20,21 In addition, we required that the ICD-9-CM diagnosis be a clinical indication for genetic testing given the age at discharge. Specifically, an age cutoff was applied based on estimates of genetic test diagnostic yield for a given ICD-9-CM in neonates versus the remaining pediatric population (Table S2). For example, a diagnosis code indicating scoliosis would only be considered suspected GD if observed in a neonate, but not in an adolescent. For each definition, we consider the background population as those patients represented by discharges with no ICD-9-CM diagnoses codes in the list.
Data analysis
Descriptive statistics, including counts, means, and standard errors, of the weighted national estimates, were used to characterize the cohort. Demographic variables were defined by HCUP.14 Due to the sampled nature of the database, reported numbers are based on discharge weights and represent national estimates.15 Analysis units are inpatient discharges, not individual patients. Year of service was 2012, and all discharges were analyzed independently. It is not possible to link multiple discharges to a single patient because records do not include patient identifiers. Total charges per discharge represent the amount billed for services by individual hospitals, exclusive of professional (physician) fees, but do not reflect actual cost of hospital services. Total charges per discharge were converted to costs using HCUP cost-to-charge ratios.16 These costs reflect actual expenses of hospital services, such as wages, supplies, and utility costs, specific to each HCUP hospital. All charges and costs are shown in 2012 US dollars.
Two-sided tests (chi-square and Fisher’s exact for categorical measures, and Student’s t tests for continuous measures) were used where appropriate with P values <0.05 considered to be statistically significant between groups. Calculations were performed separately for neonates (up to 28 days old, nonmaternal discharges) and the remaining pediatric population (29 days to less than 18 years of age at time of admission).
To account for confounding variables, e.g., region or payer type, we employed a propensity score method to compare health-care utilization metrics, accounting for systematic differences in baseline characteristics in subgroups,22,23,24,25 as utilized by other studies of KID data.26,27,28,29 Briefly, the propensity score was defined as the conditional probability of receiving a suspected GD diagnosis, using either the GD minimum or maximum classification, based on observed covariates and estimated from multivariate logistic regression. Covariates included race, sex, age, primary payer type, hospital region, the number of diagnoses and procedures included in the discharge, whether care was received at a freestanding children’s hospital, whether the birth was complicated, whether a major operating room procedure was performed, whether there was indication of a transfer into or out of the hospital, and whether there was evidence of emergency department services. The GD maximum model also included the GD minimum classification as a covariate, since the maximum estimate was inclusive of GD minimum discharges. Propensity scores were estimated using a multivariate logistic regression on GD status, with minimum and maximum definitions modeled separately. Propensity scores were trimmed to the 90th percentile because weight trimming has been shown to improve performance of propensity score weights, particularly for those estimated by logistic regression.30 Average treatment effect weights were calculated as 1/e for the treatment group and 1/(1–e) for the control group, where e is the trimmed propensity score based on GD status.31 For all health-care utilization metrics, the population average treatment effect (PATE) estimand was calculated as the difference of means between GD and the background population, after accounting for survey design, and based on the minimum or maximum GD definition. Per published recommendations,31,32 the product of the original survey weights and the propensity score weights were included in the final model estimating the PATE. This represented the average impact of having a suspected GD diagnosis on health-care utilization. All statistical analyses for weighted discharges were performed using the “survey”33 package in R.34
RESULTS
A total of 3,195,782 records, weighted to represent a national estimate of 5.85 million discharges, were identified for children less than 18 years of age in the 2012 KID. We calculated 4.37 million discharges for children less than 1 year of age, with 3.92 million of those representing neonates (nonmaternal records, 67%), with neonate status defined by admission during the first 28 days after birth. Of the neonates, a weighted 1.30 million records (33%) were labeled as complicated births.
For all analyses, unless otherwise noted, we calculated the range of potential values using the minimum and maximum estimates as defined in “Materials and Methods.” In brief, minimum estimates of GD required the discharge to have a GD-associated ICD-9-CM code as the primary diagnosis, while the maximum estimate required a GD-associated ICD-9-CM code anywhere in the diagnostic code list.
We identified a weighted estimate of 150,169 (2.6%) to 818,384 (14%) discharges with suspected GD. The range of GD estimates varied significantly by age group (Table 1). Of all the GD-linked discharges in KID, 23–45% occurred in freestanding children’s hospitals, compared with only 7.7% of all non-GD-linked discharges. Of the total number of discharges that took place in freestanding children’s hospitals (N = 577,568), 12–33% include diagnosis codes associated with GD.
Mortality rates were higher in GD-linked discharges (1.0–1.3%) compared with all other discharges (0.20%). Between 30% and 57% of suspected GD discharges also included a major operating room procedure as defined by HCUP,35 compared with 24% for all other discharges. Similarly, the percent of discharges indicating that the patient was transferred in from a different acute care hospital or other type of care facility was higher for suspected GD patients (9–15%) than all remaining patients (4%). Additional cohort demographic information is shown in Table 1 and Table S4.
GD discharges included a significantly greater number of diagnoses and procedures than non-GD discharges for both neonates and the remaining pediatric population (P < 0.0001 for both age groups, unadjusted, Table S5). For neonates, there were 3.7–6.1 more diagnoses and 1.3–4.2 more procedures in GD discharges compared with discharges with no GD-linked codes. For the remaining pediatric population, we observed a mean increase of 0.7–3.3 more diagnoses and 0.9–1.5 more procedures in GD discharges compared with discharges with no GD codes.
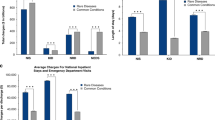
Health-care utilization was captured using length of stay (LOS), total cost, and total charges per discharge. We found significant differences in health-care utilization between discharges with and without GD-linked diagnostic codes (Table 2) among all age groups (Fig. 1). Discharges with any GD-linked diagnosis had adjusted mean total costs that were $12,017–$16,338 higher than those with no GD-linked diagnosis for the generalized pediatric population and neonates, respectively (mean total charges were $37,907–$58,511 higher). This corresponded to an adjusted mean increase in LOS of 2.4 and 6.6 days for pediatric patients and neonates, respectively. Total costs per discharge for the GD minimum estimate subgroup were on average $16,587 higher ($52,014 in mean total charges) in pediatric and $77,025 higher ($242,819 in mean total charges) in neonates than those with no GD-linked diagnosis. GD maximum estimate discharges had a mean LOS 1.8 days longer in pediatric patients and 18.6 days longer in neonates (Table 2).
GD maximum estimate discharges were aggregated according to clinical ICD-9-CM categories based on ICD-9-CM coding groups and are annotated in Table S1. Sixty-six percent of GD maximum estimate discharges had a single GD-linked ICD-9 code, and 84% had diagnosis codes in a single diagnosis group (see Figures S1 and S2). Mean total costs and charges generally increased incrementally with a larger number of GD-linked diagnoses per discharge (Fig. 2). When considering between one and seven diagnoses, which shows a linear relationship between number of codes and total charges and costs, we observed a mean incremental total cost of $13,999 (total charge of $43,862) per additional GD-linked code (Table S6). After seven diagnoses, the relationship is less clear, likely due to small sample sizes. Discharges with eight or more GD-linked codes represent only 4% of neonatal and 2% of pediatric discharges.
A subanalysis examining the confidence that a code is positively linked to genetic disease (see “Materials and methods” and Table S2) yielded results that show similar trends (Table S7). Discharges with codes most strongly linked to GD represent the smallest subpopulation (2.8% of all discharges with “definite” codes) with the highest mean health-care utilization compared with subgroups of codes that are “highly likely” or “positively associated” with GD.
The distribution of discharges and health-care utilization varied widely among diagnostic disease groupings and age group (Table 3 and Table S8). Discharges with diagnosis codes in the congenital anomalies category (ICD-9-CM codes 740.xx–759.xx) had the largest number of GD-linked discharges, as well as largest aggregate cost, in both neonates and the remaining pediatric population. Congenital anomalies accounted for an aggregate total cost of $1.4 to $8.1 billion in neonates ($4.1 to $27.6 billion, or 3.3–22% of the $124 billion aggregate total pediatric charges) and $2.3 to $5.0 billion in the remaining pediatric population ($7.1 to $15.3 billion, or 5.7–12.4% of all aggregate total pediatric charges). For neonates, diseases of the respiratory system (ICD-9-CM codes 460.xx–519.xx) had the highest mean total charge at $434,477 to $684,581 per discharge (corresponding to mean total costs of $141,198 to $235,677 and mean LOS of 35.6 to 41.3 days). In the older pediatric age group, discharges with a perinatal diagnoses code (ICD-9-CM codes 760.xx–779.xx) had the highest mean total charge, ranging from $436,934 to $437,687. This corresponded to a mean total cost of $117,719 to $123,468 and mean LOS of 41.2 to 49.7 days. All pediatric discharges with a perinatal diagnosis occurred in patients less than 1 year of age.
Discharges with GD-associated codes show significantly higher health-care utilization across all age groups compared with discharges with no GD-associated diagnoses. Adjusted mean total costs for discharges with no GD-linked codes were $2680 in neonates and $7599 in the remaining pediatric population (unadjusted values are shown in Table S9). Mean total charges for GD-associated discharges were at least 3–12 times higher in neonates, and at least 50% higher for pediatric patients, depending on the diagnosis category (Table 3 and Table S8). Overall, the aggregate total charges for all pediatric discharges in the US in 2012 were just over $124 billion, representing total costs of $37.3 billion (Table S10). While our maximum estimate of GD in neonates was only 10.2% of all neonatal discharges, aggregate costs in this population exceeded those neonatal discharges with no associated GD-linked codes ($8.7 billion versus $8.5 billion, P < 0.0001). In nonneonatal discharges with age less than 1 year, the aggregate cost of discharges with GD-associated codes was 60% higher than discharges with none (Figure S3). While the estimated genetic disease population in this data set comprises 2.6–14% of all discharges, it makes up a larger portion of the “national bill” - 11–46% of all aggregate total charges (Table 2).
DISCUSSION
This study utilized the largest all-payer database of pediatric discharges in the United States to estimate the economic burden of pediatric patients who are suspected of having a genetic disease. We found that patients with suspected GD are high utilizers of health care, on average, undergoing up to four additional procedures and staying 2 to 18 days longer in hospital, corresponding to an increase of total costs ranging from $12,000 to $77,000 per discharge compared with patients without a GD-linked diagnosis.
The selected ICD-9 codes used in our study describe clinical features that have been previously associated with genetic disorders in the literature (Table S3). The list is intended to achieve moderate to high sensitivity for inclusion of those with underlying genetic disease. Because of the current limitations of all genetic testing and the known contributions of complex or environmental causes of these clinical presentations, the specificity for GD in this population is inevitably somewhat lower. There are not standard professional society–endorsed guidelines for use of genetic testing for each of the individual selected ICD-9 codes. There are, however, large groups of patients within our study population where guidelines have endorsed genetic testing. For example, karyotype, single-gene testing, and chromosomal microarray are recommended by the American Academy of Pediatrics or the American College of Medical Genetics and Genomics for the evaluation of patients with intellectual disability, autism, and birth defects (Table S1). The diagnostic yield of genetic testing has also been documented in other neurological disorders such as epilepsy,11 with accompanying recommendation for routine testing. With respect to isolated severe birth defects, the American Heart Association has recently reinforced recommendations for genetic testing for patients with congenital heart defects, and translational research literature has documented the important role for single-gene pathogenic variants in a wide array of congenital heart defects12 (also see Table S1).
Similar population definitions have been used in previous studies. A 1997 study by Yoon et al. that reviewed pediatric hospital discharges in two states identified a similar list of 73 ICD-9-CM codes and code ranges that capture birth defects and genetic diseases across 17 disease categories. Our list (Table S2) includes all but 5 of their identified codes.36 Our code set is also inclusive of the Arth et al. codes20 associated with birth defects, and both the Arth et al. analysis and ours derived similar proportions of discharges and costs in this population despite using different data sources. Arth et al. report 556,710 patients under 18 years old using the HCUP 2013 National Inpatient Sample database, resulting in $12.4 billion in aggregate costs; while we observed that 10.3% of discharges (N = 602,028) in the database had a diagnosis code for a congenital anomaly (ICD-9-CM 740.xx–759.xx), resulting in $13.0 billion in aggregate costs in 2012. By using a more comprehensive list of GD-linked codes, we expand on these previous studies and report on an additional 216,357 discharges in 12 other disease categories, representing an aggregate cost of $4.5 billion in 2012.
Generally, patients with more indications of genetic disease, i.e., those with multiple GD-associated ICD-9 codes, showed a mean incremental total cost per discharge of $13,999, suggesting that more complex cases result in higher health-care utilization. Additionally, the majority of health-care dollars spent on GD-associated discharges tends to skew toward younger ages, as shown in Figure S3. Overall, GD-linked discharges accounted for a disproportionally high component of health-care costs for the pediatric population. This population combines patients with known underlying genetic disorders, those with genetic disorders in which the condition is either not yet diagnosed or cannot yet be recognized due to limitations of test technologies or variant interpretation, and those with either complex or purely environmental causes of their clinical presentations. This aggregated population is relevant because it represents an estimate of the population in which genetic testing might be prudently performed and whose management and therapy might be positively affected by identification of a genetic disorder.
It is possible that the estimates of health-care burden presented here are underestimates. KID is comprised of claims data for inpatient hospital stays for a single year, and although we expect the bulk of costs to be captured in an inpatient setting, charges that may occur beyond the year data were accumulated (2012) or in other hospital settings are excluded. Additionally, due to differences in state-level reporting of revenue codes, we were unable to distinguish which discharges took place in a neonatal or pediatric intensive care unit, which are generally costlier. Similarly, only bundled total charges per discharge are reported in KID, and we were unable to distinguish between individual charges within a hospital stay. By limiting our population definition to diagnosis codes in this data set, it is possible that these results may underestimate the true economic impact of genetic diseases in childhood and therefore also underestimate the implied value of genomic testing.
Results from this study have several clinical and societal implications. For example, the majority of well-established genetic diseases, neurological diseases, and birth defect–associated discharges were not for neonates (51–89%). While we are unable to longitudinally track patients in this data set, this observation may represent either a delay in diagnosis for GD patients, or a distinct group of GD diseases that present later in childhood. Because these cases occurred at older ages, they are likely to have presented outside of a typical neonatal intensive care unit. Increased physician awareness of genetic diseases is therefore necessary in a broader care setting to better recognize GD patients. In an analysis of hospital characteristics, we observed a higher proportion of GD-linked discharges in larger, high-volume urban teaching hospitals (Table S11). These hospitals are likely better equipped with the resources and expertise to handle complex medical care issues. However, we also observed a high rate of GD-associated discharges at small, nonteaching, nonchildren’s hospitals, suggesting that a substantial number of true GD cases may present in facilities not as well prepared to accurately recognize or diagnose these cases. General practitioners may miss indicators of GD, and not all physicians are familiar with the most accurate coding for rare disease.37 This lack of awareness or local expertise in different care settings or hospital types may lead to misdiagnosis and extension of diagnostic odyssey.
In utilizing this nationally representative data set, we have presented an assessment of the economic burden of pediatric GD in the United States. Given that the unit of measure in KID is inpatient discharge, not individual patient, further research, particularly cost-effectiveness studies and prospective trials, is needed to further characterize the economic burden of the diagnostic odysseys of pediatric GD patients. Evidence is accumulating that next-generation sequencing may end the diagnostic odyssey for some GD patients, and that incorporation of comprehensive testing methodologies may result in cost savings. While we were unable to directly address this question with this study, it is possible that future studies will demonstrate that early genetic testing leads to cost savings, particularly for neonates in intensive care or pediatric patients on long-term admissions.
Collectively, we observed that GD-linked discharges accounted for 2.6–14% of pediatric inpatient admissions in 2012, but made up 11% to 46% of the pediatric “national bill” based on total charges per discharge. Children who present clinically with indications of GD have a large economic impact on the national health-care system. Improved physician awareness and earlier diagnosis, particularly with comprehensive genetic testing approaches, may serve to significantly reduce costs associated with an otherwise lengthy diagnostic odyssey in pediatric GD patients.
Change history
14 February 2019
This Article was originally published under Nature Research's License to Publish, but has now been made available under a [CC BY-NC-ND 4.0] license. The PDF and HTML versions of the Article have been modified accordingly.
References
Veltman JA. Genomic microarrays in clinical diagnosis. Curr Opin Pediatr. 2006;18:598–603.
Lu X, Shaw CA, Patel A, et al. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS ONE. 2007;2:e327.
Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764.
OMIM. Baltimore, MD: Johns Hopkins University, McKusick-Nathans Institute of Genetic Medicine. 2017. https://omim.org/.
Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347.
Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879.
Taylor JC, Martin HC, Lise S, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet. 2015;47:717–726.
Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704.
Jin SC, Homsy J, Zaidi S, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–1601.
Manning M, Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–745.
Olson H, Shen Y, Avallone J, et al. Copy number variation plays an important role in clinical epilepsy. Ann Neurol. 2014;75:943–958.
Sifrim A, Hitz MP, Wilsdon A, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–1065.
Healthcare Cost and Utilization Project (HCUP). The Healthcare Cost and Utilization Project fact sheet. Agency for Healthcare Research and Quality. 2017. https://www.hcup-us.ahrq.gov/news/exhibit_booth/hcup_fact_sheet.jsp. Accessed 20 August 2017.
Healthcare Cost and Utilization Project (HCUP). Introduction to the HCUP KIDS’ Inpatient Database (KID): 2012. Agency for Healthcare Research and Quality. 2015. https://www.hcup-us.ahrq.gov/db/nation/kid/kid_2012_introduction.jsp. Accessed 20 August 2017.
Healthcare Cost and Utilization Project (HCUP). Producing national HCUP estimates—accessible version. Agency for Healthcare Research and Quality. 2015. http://www.hcup-us.ahrq.gov/tech_assist/nationalestimates/508_course/508course.jsp. Accessed 20 August 2017.
Healthcare Cost and Utilization Project (HCUP). Cost-to-charge ratio files: 2012 Kids’ Inpatient Database (KID) user guide. Agency for Healthcare Research and Quality. 2014. https://www.hcup-us.ahrq.gov/db/state/CCR2012KIDUserGuide.pdf. Accessed 21 August 2017.
Köhler S, Vasilevsky NA, Engelstad M, Foster E, et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 2017;45:D865–D876.
Healthcare Cost and Utilization Project (HCUP). HCUP NEDS description of data elements. Agency for Healthcare Research and Quality. 2015. https://www.hcup-us.ahrq.gov/db/vars/dxn/nedsnote.jsp. Accessed 21 August 2017.
Centers for Medicare and Medicaid Services and the National Center for Health Statistics. ICD-9-CM official guidelines for coding and reporting. https://www.cdc.gov/nchs/data/icd/icd9cm_guidelines_2011.pdf. Accessed 19 August 2017.
Arth AC, Tinker SC, Simeone RM, Ailes EC, Cragan JD, Grosse SD. Inpatient hospitalization costs associated with birth defects among persons of all ages—United States, 2013. MMWR Morb Mortal Wkly Rep. 2017;66:41–46.
Moffitt KB, Case AP, Farag NH, Canfield MA. Hospitalization charges for children with birth defects in Texas, 2001 to 2010. Birth Defects Res A Clin Mol Teratol. 2016;106:155–163.
Gagne JJ, Thompson L, O’Keefe K, Kesselheim AS. Innovative research methods for studying treatments for rare diseases: methodological review. BMJ. 2014;349:g6802–g6802.
Austin PC. An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivariate Behav Res. 2011;46:399–424.
Little RJ, Rubin DB. Causal effects in clinical and epidemiological studies via potential outcomes: concepts and analytical approaches. Annu Rev Public Health. 2000;21:121–145.
Brookhart MA, Wyss R, Layton JB, Stürmer T. Propensity score methods for confounding control in nonexperimental research. Circ Cardiovasc Qual Outcomes. 2013;6:604–611.
Nylund CM, Goudie A, Garza JM, Fairbrother G, Cohen MB. Clostridium difficile infection in hospitalized children in the United States. Arch Pediatr Adolesc Med. 2011;165:451–457.
Gao Y, Pugely A, Karam M, Phisitkul P, Mendoza S, Johnston RC. Is hospital teaching status a key factor in hospital charge for children with hip fractures?: preliminary findings from KID database. Iowa Orthop J. 2013;33:130–135.
Adams DJ, Eberly MD, Goudie A, Nylund CM. Rising vancomycin-resistant Enterococcus infections in hospitalized children in the United States. Hosp Pediatr. 2016;6:404–411.
Bekelis K, Connolly ID, Do HM, Choudhri O. Operative volume and outcomes of cerebrovascular neurosurgery in children. J Neurosurg Pediatr. 2016;18:623–628.
Lee BK, Lessler J, Stuart EA. Weight trimming and propensity score weighting. PLoS One. 2011;6:e18174.
DuGoff EH, Schuler M, Stuart EA. Generalizing observational study results: applying propensity score methods to complex surveys. Health Serv Res. 2014;49:284–303.
Schuler M, DuGoff EH, Stuart EA. Generalizing observational study results: applying propensity score methods to complex surveys. Oral presentation at: National Conference on Health Statistics; August, 2012; Washington, DC. https://www.cdc.gov/nchs/ppt/nchs2012/ss-33_schuler.pdf.
Lumley T Survey: analysis of complex survey samples [computer program]. Release R package version 3.32. Vienna, Austria: R Foundation for Statistical Computing, 2016.
R Core Team. R: A language and environment for statistical computing [computer program]. Release 3.3.2. Vienna, Austria: R Foundation for Statistical Computing, 2016.
Healthcare Cost and Utilization Project (HCUP). HCUP KID description of data elements. Agency for Healthcare Research and Quality. 2016. http://www.hcup-us.ahrq.gov/db/vars/orproc/kidnote.jsp. Accessed 1 September 2017.
Yoon PW, Olney RS, Khoury MJ, Sappenfield WM, Chavez GF, Taylor D. Contribution of birth defects and genetic diseases to pediatric hospitalizations. A population-based study. Arch Pediatr Adolesc Med. 1997;151:1096–1103.
Walker CE, Mahede T, Davis G, et al. The collective impact of rare diseases in Western Australia: an estimate using a population-based cohort. Genet Med. 2017;19:546–552.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
All authors are employees of Illumina, Inc.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gonzaludo, N., Belmont, J.W., Gainullin, V.G. et al. Estimating the burden and economic impact of pediatric genetic disease. Genet Med 21, 1781–1789 (2019). https://doi.org/10.1038/s41436-018-0398-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0398-5
Keywords
This article is cited by
-
The health-care utilization and economic burden in patients with genetic skeletal disorders
Orphanet Journal of Rare Diseases (2024)
-
Rapid genomic sequencing for genetic disease diagnosis and therapy in intensive care units: a review
npj Genomic Medicine (2024)
-
Physicians’ use and perceptions of genetic testing for rare diseases in China: a nationwide cross-sectional study
Orphanet Journal of Rare Diseases (2023)
-
Rare diseases, common barriers: disparities in pediatric clinical genetics outcomes
Pediatric Research (2023)
-
Bayesian cost-effectiveness analysis of Whole genome sequencing versus Whole exome sequencing in a pediatric population with suspected genetic disorders
The European Journal of Health Economics (2023)