Abstract
Purpose
The nature of phenylalanine hydroxylase (PAH) variants determines residual enzyme activity, which modifies the clinical phenotype in phenylketonuria (PKU). We exploited the statistical power of a large genotype database to determine the relationship between genotype and phenotype in PKU.
Methods
A total of 9336 PKU patients with 2589 different genotypes, carrying 588 variants, were investigated using an allelic phenotype value (APV) algorithm.
Results
We identified 251 0-variants encoding inactive PAH, and assigned APVs (0 = classic PKU; 5 = mild PKU; 10 = mild hyperphenylalaninaemia) to 88 variants in PAH-functional hemizygous patients. The genotypic phenotype values (GPVs) were set equal to the higher-APV allele, which was assumed to be dominant over the lower-APV allele and to determine the metabolic phenotype. GPVs for 8872 patients resulted in cut-off ranges of 0.0–2.7 for classic PKU, 2.8–6.6 for mild PKU and 6.7–10.0 for mild hyperphenylalaninaemia. Genotype-based phenotype prediction was 99.2% for classic PKU, 46.2% for mild PKU and 89.5% for mild hyperphenylalaninaemia. The relationships between known pretreatment blood phenylalanine levels and GPVs (n = 4217), as well as tetrahydrobiopterin responsiveness and GPVs (n = 3488), were significant (both P < 0.001).
Conclusions
APV and GPV are powerful tools to investigate genotype–phenotype associations, and can be used for genetic counselling of PKU families.
Similar content being viewed by others
INTRODUCTION
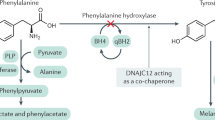
Phenylketonuria (PKU; MIM 261600) is a heterogeneous group of diseases caused by variants in the phenylalanine hydroxylase (PAH) gene.1 PAH catalyses the oxidation of phenylalanine (Phe) to tyrosine.2 The cofactor tetrahydrobiopterin (BH4), iron and molecular oxygen are required for its full activity.3 PAH deficiency presents phenotypically as mild hyperphenylalaninaemia (MHP; blood Phe = 120–600 µmol l–1), mild PKU (mPKU; blood Phe = 600–1200 µmol l–1) or classic PKU (cPKU; blood Phe > 1200 µmol l–1), with mPKU and cPKU requiring treatment. Inheritance is autosomal recessive with an average prevalence of 1:10,000.4 Treatment with a low-Phe diet enables near-normal cognitive development in affected children.5 Alternative treatment options include pharmacological supplementation with BH4, which acts as a chaperone for PAH, and enzyme substitution with phenylalanine ammonia lyase (in phase 3 development).6
More than 1000 PAH variants have been reported to the locus-specific database PAHvdb (http://www.biopku.org/home/pah.asp): 65% are in-frame amino acid substitutions, including changes to nonsense codons, and other common variants, such as deletions (15%), splice variants (12%), insertions, duplications or indels (4%), and synonymous variants (4%), indicating that PKU is a highly heterogeneous disease.7,8 Relationships between genotypes and metabolic phenotypes reported in the past have varied widely, from no correlation to incomplete or substantial correlation.9,10 Associations between genotypes and in vitro residual PAH activity have been documented for several PAH variants.11,12,13,14 Epigenetic factors (e.g., DNA methylation) and gene products involved in protein stability (e.g., chaperones and proteolytic enzymes), as well as interallelic complementation (positive or negative), are also factors influencing the phenotype.15
Many of the missense PAH variants that retain substantial residual PAH activity are associated with BH4 responsiveness.16 Patients with BH4-responsive PKU may benefit from oral administration of synthetic BH4 (sapropterin dihydrochloride), which lowers blood Phe levels.17,18 Hence, genotype analysis has the potential to identify likely BH4 responders.19,20
Prediction of metabolic phenotypes by genotype in PKU was proposed by Guldberg et al.21 on the basis of 105 PAH variants in 297 functionally hemizygous patients. Some of the variants were reported only once or twice. The PAH variants were assigned arbitrary values (AVs) based on the power of 2: 1 = cPKU; 2 = moderate PKU; 4 = mPKU; and 8 = MHP. Although the genotype is characterized by the sum of two AVs, due to the power scale, the proposed phenotypic effect is based on the higher AV. This AV system has since been used in several studies, where genotype-based phenotype predictions were correct in approximately 54% of patients; however, numbers of patients were always limited.22,23
In this study, we investigated genotype–phenotype associations in 9336 PKU patients with various degrees of hyperphenylalaninaemia and with a known genotype. We calculated allelic phenotype values (APVs) based on the frequencies of the metabolic phenotype (i.e., cPKU, mPKU or MHP) for genotypes presenting in a functionally hemizygous state. A scale for APV, ranging from 0 for cPKU to 5 for mPKU and 10 for MHP, was introduced. Genotypic phenotype values (GPVs) were calculated for all patients and compared with blood Phe levels and BH4 responsiveness.
MATERIALS AND METHODS
Patients, phenotypes and genotypes
A total of 9336 PAH-deficient patients with a known genotype had been tabulated in the BIOPKU database (http://www.biopku.org/home/biopku.asp) as of October 2017. Individual information was collected from the published literature or records submitted online. In addition to the genotype, BIOPKU includes information on the phenotype, BH4 responsiveness (following a BH4 challenge) and blood Phe concentrations (where reported). Based on the highest reported pretreatment blood Phe level, patients’ phenotypes were classified as cPKU, mPKU or MHP. However, neither the method used for blood Phe quantification nor the age at measurement was reported. Similarly, the protocols for the BH4 challenge were different, and only patients who responded to oral BH4 administration with a decrease in blood Phe by at least 30% within 24–48 h were designated as responders. BH4 responsiveness was reported as “yes” or “no”. In total, 1373 (53%) of the 2589 different genotypes in the BIOPKU databases were reported as present in only a single patient. Data in the BIOPKU are anonymized and cannot be traced back to the families.
The BIOPKU database is linked to the PAHvdb database, containing a total of 1041 PAH variants as of October 2017.8 PAHvdb follows the Human Genome Variation Society (http://www.hgvs.org/content/guidelines) nomenclature recommendations.
Patients in the BIOPKU database harboured 588 different PAH variants, 247 of which were initially defined as 0-variants, which have presumed or confirmed zero PAH activity and are thus associated with cPKU (Supplementary Table 1). Most 0-variants were: base substitutions, deletions or insertions that introduced a premature stop codon with or without a frame-shift; splice site variants that completely impaired correct messenger RNA (mRNA) splicing; or large deletions. There were also some missense variants with no in vitro enzyme activity. All 0-variants were tested against each other to confirm a cPKU-only set.
Two-stage algorithm for validation of APVs
The aim of the algorithm was to assign APVs to variants, which were then used to calculate GPVs for the prediction of metabolic phenotypes and to test the APV model. APVs range between 0 and 10, with APV = 0 indicating cPKU, APV = 5 mPKU and APV = 10 MHP. Three basic assumptions were incorporated into the model: (1) the APV of a so-far unknown variant present in a functionally hemizygous constellation (i.e., with another known 0-variant) corresponds to the phenotype of this constellation; (2) each variant belongs to one of three phenotypes (cPKU, mPKU or MHP); and (3) the observed metabolic phenotype is the result of the highest PAH activity of the two variants (i.e., the model does not take into account potential interallelic complementation (positive or negative)).
In the BIOPKU database, each variant was classified as either a 0-variant with no residual activity or a non-0-variant with presumed or observed residual activity. Initially, all non-0-variants were assigned as 'unknown'. In stage 1, variants with frequencies ≥9 and classified as belonging to the same phenotype group in at least 90% of the cases were assigned APV values of 0, 5 or 10 and considered to be strong predictors (for details, see Supplementary Figure 1). Cases not congruent with the at least 90% concordant phenotypes were classified as errors and excluded from further analysis. The same procedure was applied to variants occurring in at least 4 but in fewer than 9 patients (4 ≤ n < 9), with the same phenotype leading to weak predictors. Table 1 summarizes the logic of predicting phenotypes from two alleles with known APVs. In stage 2, for all remaining unknown variants not fulfilling the frequency and percentage of concordance criteria, APVs were computed using the following equation:
where % indicates the percentage of phenotypes for a given functionally hemizygous genotype. For example, for a variant occurring in 10 functionally hemizygous patients once as cPKU (1%), 7 times as mPKU (7%) and twice as MHP (2%), the APV = ((1 × 0) + (7 × 5) + (2 × 10))/100 = 5.5, indicating mPKU.
All APVs calculated from functionally hemizygous cases were compared with those calculated from homozygous cases when n ≥ 9 (strong predictors). APVs were also calculated using the same equation for variants that occurred in fewer than 9 patients (4 ≤ n < 9) and were designated weak predictors. APVs when n < 4 and more than 1 phenotype class is predicted are not informative and should be used with caution.
GPVs
GPVs combine information from both alleles and indicate the metabolic phenotype. The GPV was calculated as APVmax (the APV with the higher value of the two APVs) because the sum of the two APVs would not always distinguish between different phenotypes. For example, using the sum of APVs, a genotype with APV1 = 5 and APV2 = 5 (both associated with mPKU) and a genotype with APV1 = 0 (cPKU) and APV2 = 10 (MHP) would both result in a GPV of 10, but they would probably not express the same phenotype.
GPV, blood Phe levels and responsiveness to BH4
GPVs were compared with mean blood Phe levels using the nonlinear regression equation (2) for all cases in which Phe was reported (see “Patients, phenotypes and genotypes”):
where GPV is equal to APVmax, a and b are constants, e is Euler's number and ε is an error term.
GPV and responsiveness to BH4
GPVs were evaluated with regards to BH4 responsiveness in all cases where BH4 challenges were reported.
Statistical analyses
All statistical analyses were performed with the R language for statistical computing and graphics.24 Cut-off values for predicting the metabolic phenotype from the GPV were determined by linear discriminant analysis.25 Analysis of variance (ANOVA) was used to evaluate relationships among several nominal predictor variables and a continuous response variable. ANOVA post hoc comparisons used estimated marginal means.26 The Mann-Whitney U-test was used to compare two groups, with a Bonferroni P value adjustment when necessary. Receiver operating characteristic curves relating the sensitivity and specificity of GPVs and AVs for the three phenotypes were calculated using the R package pROC27 and compared with DeLong’s test.
RESULTS
Validation of 0-variants
Initially, APV = 0 was assigned to 247 variants (Supplementary Table 1). Out of 3283 cases, only 19 with 2 0-variants were not reported as cPKU. The expert-ranked 0-variants c.1315 + 6 T > A/p.?(IVS12 + 6 T > A), c.510–21_665del177/p.?(IVS5–21del177) and c.1099dupC/p.L367Pfs*27 were re-classified as unknown (Supplementary Table 1) because c.1315 + 6 T > A and c.510–21_665del177 occurred once each with another 0-variant, resulting in mPKU, and c.1099dupC occurred twice with another 0-variant (once with a cPKU phenotype and once with an mPKU phenotype). The remaining 16 cases with an mPKU phenotype were judged to be misclassified and excluded from further analysis.
Genotypes consisting of APV = 0 and 'unknown' were combined to identify new 0-variants. In iteration 1, 6 unknown variants were assigned APV = 0 (c.473 G > A/p.R158Q, c.728 G > A/p.R243Q, c.809 G > A/p.R270K, c.847 A > T/p.I283F, c.969 + 5 G > A/p.?(IVS9 + 5 G > A) and c.1066–3 C > T/p.?(IVS10–3C > T)). In iteration 2, c.1183 G > C/p.A395P was identified as a further 0-variant (Supplementary Table 1).
Validation of MHP and mPKU variants
Genotypes with APV = 0 plus 'unknown' were tabulated to identify MHP variants (APV = 10). In the first iteration, eight MHP variants were identified (c.965 C > G/p.A322G, c.1243 G > A/p.D415N, c.527 G > T/p.R176L, c.890 G > A/p.R297H, c.259 A > C/p.S87R, c.1139 C > T/p.T380M, c.688 G > A/p.V230I and c.734 T > C/p.V245A; Table 2). Further iterations did not reveal new MHP variants.
Genotypes with APV = 0 and 'unknown' were then tabulated to identify variants with APV = 5. In the first iteration, five variants were identified (c.386 A > G/p.D129G, c.722 G > A/p.R241H, c.204 A > T/p.R68S, c.1241 A > G/p.Y414C and c.1012 G > T/p.D338Y; Table 2). Further iterations did not reveal new variants.
Genotypes with APV = 5 and 'unknown' were tabulated to identify additional MHP variants. No additional variants could be assigned.
Assignment of APV to unknown variants
APVs were assigned to 33 frequent (n ≥ 9) and 32 less frequent (4 ≤ n < 9) variants (Table 2). Ten variants occurring in only three patients each but with a uniform phenotype were included as well. All other variants were assigned as unknown and given no APV, although this information may change as the numbers of reported patients increase.
Effect of homozygous genotypes on APVs
From the 30 most frequent homozygous genotypes, all of which occurred in at least 9 cases (Supplementary Table 2), 20 did not differ from the corresponding functionally hemizygous genotypes; however, 19 out of these were cPKU. In 5 homozygous genotypes, the GPV was higher, but only by ≤1.0. The following variants had the largest discrepancies in APV (2.4–3.4 units): c.194 T > C/p.I65T, c.782 G > A/p.R261Q and c.721 C > T/p.R241C. For the c.782 G > A/p.R261Q and c.194 T > C/p.I65T variants, APVs were between the cut-offs for cPKU and mPKU, while for c.721 C > T/p.R241C, the APV was between mPKU and MHP. The most striking difference in APVs was found for the splice site variant c.1066–3 C > T/IVS10–3C > T, which is associated with cPKU in the functionally hemizygous state, but also reliably reported in three homozygous MHP patients from Turkey and Spain.
Association between GPV and phenotypes and the definition of cut-off values
GPV was compared among the phenotypes of patients in the BIOPKU database (n = 8673), with 5146 (59.4%) reported as cPKU, 2101 (24.2%) as mPKU and 1423 (16.4%) as MHP (Figure 1a). These numbers are probably representative of the distribution of metabolic phenotypes of the global PKU population. The GPV cut-off ranges calculated by linear discriminant analysis were 0.0–2.7 for cPKU, 2.8–6.6 for mPKU and 6.7–10.0 for MHP, where 5107/5149 (99.2%) cases were in the range defined for cPKU and only 42/5149 (0.8%) were above the upper cut-off value (discriminant coefficients are available in Supplementary Materials 2). Within the cut-off range for mPKU were 970/2101 (46.2%) cases, with 852/2101 (40.6%) below and 277/2101 (13.2%) above the range. For MHP, 1273/1423 (89.5%) cases were within the cut-off range, and 150/1423 (10.5%) fell below.
a Boxplots (median, 25th–75th percentile, ±1.5) of the genotypic phenotype value (GPV; that is, the maximum allelic phenotype value of the two genotype-determining alleles) for three metabolic phenotypes in 8669 phenylketonuria (PKU) patients. Circles represent outliers. Numbers show the numbers of cases, with the number of outliers in brackets. Dark grey areas represent cut-off ranges. cPKU classic PKU, mPKU mild PKU, MHP mild hyperphenylalaninaemia. b Boxplots (median, 25th–75th percentile, ±1.5) for GPV in 3665 tetrahydrobiopterin (BH4) responders and non-responders. Only cases tested for BH4 responsiveness were included. Circles represent outliers. The number of cases is shown below the boxes, with the number of outliers in brackets. For a definition of BH4 responsiveness, see “Patients, phenotypes and genotypes”
Comparison between GPV and BH4 responsiveness
GPV and BH4 responsiveness data were available for 3665 patients (Figure 1b). For 2133 non-responders, the GPV (median (25th–75th percentile)) was 0 (0–5); 193 patients with a GPV >4.0 were defined as statistical outliers. There was no outlier in the 1532 BH4 responders with a GPV (median (25th–75th percentile)) of 5.0 (2.4–8.9) (P < 0.0001, Mann-Whitney U-test).
Finally, a two-way ANOVA of GPV, with BH4 responder status (yes or no) and metabolic phenotype (cPKU, mPKU or MHP) as independent variables revealed a significant interaction (F2,3659 = 3.32, P = 0.036). Post hoc comparisons showed that mean GPVs were significantly different between all combinations of metabolic phenotypes and BH4 responder status except for MHP, where the difference between GPVs for responders versus non-responders was not significant. However, it should be noted that in the BIOPKU dataset, 103 cPKU patients were classified as BH4 responders—a result that requires explanation.
Cross-validation of GPVs and Guldberg’s AVs
Guldberg’s AVs and GPVs were cross-validated using the genotype and phenotype information from 358 Spanish patients (128 cPKU, 80 mPKU and 150 MHP)19 not included in the BIOPKU dataset. For all patients, APVs (and thus GPVs and AVs) that were previously reported by Guldberg et al.21 were known for both alleles (Supplementary Figure 2a, b). The GPV separated cPKU (median (25th–75th percentile) = 0 (0–1.5)) from MHP (median (25th–75th percentile) = 9.1 (8.9–10.0)), while mPKU (median (25th–75th percentile) = 1.8 (1.3–5.0)) was partially overlapping with cPKU. Descriptive analysis of AV revealed 2 (2–5) (median (25th–75th percentile)) for cPKU, 5 (5–8) for mPKU and 9 (9–12) for MPH (P values, two tailed: always <0.0001, U-test, Bonferroni adjusted) (Supplementary Figure 2a, b).
Following Guldberg’s genotype–phenotype classification,21 genotypes with AV = 2 were classified as cPKU, AVs 3–8 were classified as mPKU and AVs 9–16 were classified as MHP. The sensitivity and specificity of predictions by GPVs versus the AV system were analysed with receiver operating characteristic curves (Supplementary Figure 3). Although in absolute measures the differences were small, the areas under the curve (AUCs) were significantly larger for cPKU (AUC for APV = 89.96; AUC for AV = 87.89; DeLong’s z = −2.04, P = 0.042) and MHP (AUC for APV = 97.35; AUC for AV = 94.02; DeLong’s z = 3.05, P < 0.01) for GPV, indicating higher sensitivity and specificity for GPV. AUCs for mPKU (AUC for APV = 61.58; AUC for AV = 59.76; DeLong’s z = −1.3, P = 0.19) were not significantly different. For cPKU, the positive predictive value (PPV) for GPV was 0.7, compared with 0.8 for AV; however, the sensitivity of GPV was much higher (0.98) for GPV than AV (0.58). This discrepancy in sensitivity corresponds to a false positive rate of 0.42. Predictions of mPKU based on APV and AV were not only similar, but also their PPVs were relatively low for GPV (0.6) as well as AV (0.4). Predictions based on GPV showed higher PPVs (1.0), sensitivity (0.9) and specificity (1.0) than predictions based on AV (0.99, 0.8 and 0.99, respectively) (Supplementary Table 3).
Alignment of the APV scale to blood Phe values
To compare APVs with the maximum untreated Phe value, we calculated medians and 25th–75th percentiles for data provided in the entire BIOPKU database. These were 1677 μmol l–1 (1116–3919 μmol l–1) for cPKU (n = 5096), 835 μmol l–1 (506–1220 μmol l–1) for mPKU (n = 1991) and 341 μmol l–1 (91–623 μmol l–1) for MHP (n = 1331). The overlapping areas between the three phenotypes and the upper open area for blood Phe in cPKU indicate where misclassification of phenotypes may occur. The alignment of APVs to blood Phe is shown in Figure 2.
a–c Alignment of the allelic phenotype value (APV) scale (a) to three phenotype classes (b) and the pretreatment blood Phe levels (median, 5th–95th percentile) of 8418 phenylketonuria (PKU) patients tabulated in the BIOPKU database (c). Note the overlapping areas in blood Phe levels between mild hyperphenylalaninaemia (MHP) and mild PKU (mPKU), and between mPKU and classic PKU (cPKU)
Association between GPV and blood Phe
The association of GPVs with pretreatment blood Phe values in 4217 of the patients is shown in Figure 3a, b.
a Scatterplot of pretreatment blood phenylalanine (Phe) levels of 4435 phenylketonuria (PKU) patients as a function of the genotypic phenotype value (GPV) in groups separated by metabolic phenotype. The dashed orange line shows the nonlinear regression model (see main text). cPKU classic PKU, mPKU mild PKU, MHP mild hyperphenylalaninaemia. b Contour plot of two-dimensional densities of pretreatment levels of Phe and GPV. Darker regions indicate higher frequencies of pairs of GPV and pretreatment Phe levels
For this subsample, Phe levels (mean; SD; n) were: 1737 µmol l–1; 633 µmol l–1; 2206 for cPKU; 828 µmol l–1; 223 µmol l–1; 1166 for mPKU; and 358 µmol l–1; 136 µmol l–1; 845 for MHP. GPVs (mean; SD; n) were highest in MHP (8.7; 1.44; 845), mid-range in mPKU (4.10; 2.33; 1166) and lowest in cPKU (0.47; 0.47; 2206). Nonlinear regression analysis identified a significant relationship between blood Phe and GPV: significant coefficients a = 1763 and b = −0.17 (both P < 0.001) indicate that with increasing GPV, pretreatment blood Phe decreased significantly (Figure 3a). A contour graphic of the two-dimensional density of pretreatment blood Phe and GPVs shows the expected high negative association in all phenotypes (Figure 3b).
Online access to APVs and GPVs
APVs are tabulated in the PAHvdb locus-specific database (http://www.biopku.org/pah) and linked with the BIOPKU genotypes database (http://www.biopku.org/biopku). APVs were dynamically calculated from the actual BIOPKU dataset. Following a search for a specific genotype in the BIOPKU database, it reports the corresponding GPV and predicted phenotype. A detailed walkthrough of the BIOPKU application is presented in Supplementary Materials 1.
DISCUSSION
Understanding the genetic basis of inherited metabolic diseases is an ongoing challenge. Multi-omics systems have increased the amount of genomic (e.g., single-nucleotide polymorphisms, copy number variations, loss of heterozygosity and genomic rearrangements), epigenomic (e.g., DNA methylation, histone modification, chromatin accessibility, transcription factor binding and microRNA), transcriptomic (e.g., gene expression and alternative splicing), proteomic (e.g., protein expression and post-translational modifications) and metabolomic (e.g., metabolite profiling in plasma, urine or cerebrospinal fluid) data flowing into the databases of genetic information available for analysing the phenome.28 Despite the increasing amount of data being generated, the association between multi-omics and phenomics is not always predictable. In many genetic diseases, the small numbers of reported cases limit our ability to perform feasible genotype–phenotype correlations, and proteomic and epigenomic constellations complicate data interpretation. For this study, to explore correlations in the PKU population, we used information from a large database of over 9000 patients to establish a relationship between gene variants and metabolic phenotypes.
Our approach differs from those obtained by previous simple correlations derived from a limited number of genotypes and metabolic phenotypes29,30,31 or between genotypes and BH4 responsiveness.19,20,32 While these studies indicate that there are no simple genotype–phenotype correlations, in many cases, a reasonable match (60–70%) has been observed between predicted and observed phenotypes. For example, genotype was informative for BH4 responsiveness in approximately 25% of cases.20,31 We found a significant (P < 0.0001) association between pretreatment blood levels and GPV in a group of 4435 patients containing all 3 phenotype classes. As expected, higher GPVs were significantly correlated with lower blood Phe levels. No outliers were detected within the group of 1532 BH4 responders, and the two groups were well separated on the GPV scale (P < 0.0001) (Figure 1b). We found that BH4 responsiveness was clearly less frequent in more severe variants (APV < 2) and more common in those with APV > 5. Out of 2133 non-responders, 193 were found to have GPVs broadly ranging between 4 and 10, perhaps because of the non-standardized protocols used for BH4 challenge.33
Another approach to correlate genotypes with phenotypes is to analyse the transient expression of PAH variants in eukaryotic cell systems and measure their in vitro residual activity. In early studies, expression analyses of PAH variants showed that residual enzyme activity was strongly correlated with pretreatment blood Phe levels and Phe tolerance.11,29,34 However, the amount of mutant PAH protein and residual activity could be modulated by the in vitro experimental conditions, and the observed in vitro metabolic variation could, therefore, be explained by inter-individual variation in protein quality control systems (e.g., chaperones and proteases).13 Similar inconsistencies between genotypes and metabolic phenotypes have frequently been reported for all three PKU classes35,36, and the co-expression of two distinct PAH variants suggests that a possible dominance effect (positive or negative) by one of the variants on residual PAH activity may be the result of interallelic complementation.15 In our APV model, no such events appeared relevant.
If residual enzymatic function is the most determining factor for a clinical phenotype, identical PAH variants should cause the same effects on PAH function and therefore the same clinical phenotype. Several potential mechanisms may account for the presence of discordant cases. Variations in the fidelity of splicing mechanisms can result in multiple mRNAs, possibly including some normal mRNA transcripts resulting in enzyme activity. Because the mechanisms responsible for splicing are independently inherited from PAH, the amount of normal mRNA encoding PAH may vary among individuals carrying the same splicing variant. This mechanism may explain why discordant phenotypes are commonly associated with splicing variants. One example from our study is the c.1066–3 C > T splicing variant, which has a variable phenotype. This variant was found to be severe in a functionally hemizygous state, but was also reported and confirmed in three homozygous MHP patients. These patients represent an example of phenotypic variation associated with a splicing variant, and indicate one of the challenges faced when classifying a continuously graded spectrum of clinical phenotypes into distinct categories.
Structural instability and misfolding of the PAH protein may also alter enzyme activity independent of the associated variant.37 Since mechanisms of degradation and synthesis are independently inherited from PAH, different individuals with the same PAH variant may have different steady-state levels of PAH activity. Molecular chaperones, including heat shock proteins (HSPs), are essential components of the protein quality control system. The 70 kDa HSPs (HSP70s) are versatile chaperones that maintain protein homoeostasis (proteostasis) by controlling the proper folding, degradation and translocation of many client proteins or substrates. HSP40s, or J-proteins, such as DNAJC12, function as co-chaperones of HSP70s. DNAJC12 has been shown to interact with PAH and DNAJC12 variants to cause hyperphenylalaninaemia if mutated. DNAJC12 is responsible for the proper folding and intracellular stability of PAH.38 Although this is rather speculative, individuals with the same PAH genotype may have DNAJC12 variants affecting PAH folding, ubiquitin-tagged degradation and enzyme activity, which would result in different phenotypes.
A wide range of genotype–phenotype associations present with well-established properties that appear to be common to all of them. These include inconsistency in genotype, meaning that some genotypes map to different phenotypes, as found in PKU. Aside from the aforementioned epigenetic factors, interallelic complementation and regulatory factors, such as chaperones, the quality of information and the number of cases per genotype are essential to evaluate the power of a genotype-based phenotypic prediction. Misclassification of phenotypes is the main reason for differences between studies.39 We took advantage of a large database that contains the genotypes and phenotypes of 9336 PKU patients from all over the world, 3665 of whom were challenged with BH4, to establish a model to quantify allelic severity according to APVs and predict phenotypes based on genotypes. As previously proposed,21 one assumption was essential to characterize the observed genotypes: a variant with a higher APV was always dominant over one with a lower APV (severe). Thus, it was not the sum of both alleles (e.g., APV1 + APV2), but rather the APVmax that determined the metabolic phenotype. APV values were added to the PAHvdb locus-specific database, which is linked to the BIOPKU genotypes database and calculates the GPV for a selected genotype. The addition of new records to the BIOPKU database may alter the APV slightly, and if the number of cases reaches ≥9, the APV may change from a weak predictor to a strong one.
We hope that the APV/GPV model presented here and its implementation into BIOPKU will help professionals develop evidence-based metabolic phenotyping, particularly in instances when treatment recommendations are unclear (e.g., due to borderline blood Phe levels), and improve genetic counselling of patients’ families. It can also serve as a model for how large genotype–phenotype databases with well-defined phenotype groups can contribute to the rapidly growing field of omics.40
REFERENCES
Blau N, Van Spronsen FJ, Levy HL Phenylketonuria. Lancet 2010;376:1417–1427.
Kaufman S The phenylalanine hydroxylating system. Adv Enzymol Relat Areas Mol Biol 1993;67:77–264.
Werner ER, Blau N, Thöny B Tetrahydrobiopterin: biochemistry and pathophysiology. Biochem J 2011;438:397–414.
Hardelid P, Cortina-Borja M, Munro A et al. The birth prevalence of PKU in populations of European, South Asian and sub-Saharan African ancestry living in South East England. Ann Hum Genet 2008;72:65–71.
Vockley J, Andersson HC, Antshel KM et al. Phenylalanine hydroxylase deficiency: diagnosis and management guideline. Genet Med 2014;16:188–200.
Blau N, Longo N Alternative therapies to address the unmet medical needs of patients with phenylketonuria. Expert Opin Pharmacother 2015;16:1–10.
Blau N, Shen N, Carducci C Molecular genetics and diagnosis of phenylketonuria: state of the art. Expert Rev Mol Diagn 2014;14:655–671.
Blau N Genetics of phenylketonuria: then and now. Hum Mutat 2016;37:508–515.
Desviat LR, Perez B, Gamez A et al. Genetic and phenotypic aspects of phenylalanine hydroxylase deficiency in Spain: molecular survey by regions. Eur J Hum Genet 1999;7:386–392.
Daniele A, Scala I, Cardillo G et al. Functional and structural characterization of novel mutations and genotype–phenotype correlation in 51 phenylalanine hydroxylase deficient families from Southern Italy. FEBS J 2009;276:2048–2059.
Okano Y, Eisensmith RC, Güttler F et al. Molecular basis of phenotypic heterogeneity in phenylketonuria. N Engl J Med 1991;324:1232–1238.
Waters PJ, Parniak MA, Nowacki P et al. In vitro expression analysis of mutations in phenylalanine hydroxylase: linking genotype to phenotype and structure to function. Hum Mutat 1998;11:4–17.
Pey AL, Desviat LR, Gamez A et al. Phenylketonuria: genotype–phenotype correlations based on expression analysis of structural and functional mutations in PAH. Hum Mutat 2003;21:370–378.
Wettstein S, Underhaug J, Perez B et al. Linking genotypes database with locus-specific database and genotype–phenotype correlation in phenylketonuria. Eur J Hum Genet 2015;23:302–309.
Shen N, Heintz C, Thiel C et al. Co-expression of phenylalanine hydroxylase variants and effects of interallelic complementation on in vitro enzyme activity and genotype–phenotype correlation. Mol Genet Metab 2015;117:328–335.
Zurflüh MR, Zschocke J, Lindner M et al. Molecular genetics of tetrahydrobiopterin responsive phenylalanine hydroxylase deficiency. Hum Mutat 2008;29:167–175.
Muntau AC, Röschinger W, Habich M et al. Tetrahydrobiopterin as an alternative treatment for mild phenylketonuria. N Engl J Med 2002;347:2122–2132.
Keil S, Anjema K, van Spronsen FJ et al. Long-term follow-up and outcome of phenylketonuria patients on sapropterin: a retrospective study. Pediatrics 2013;131:1881–1888.
Aldamiz-Echevarria L, Llarena M, Bueno MA et al. Molecular epidemiology, genotype–phenotype correlation and BH4 responsiveness in Spanish patients with phenylketonuria. J Hum Genet 2016;61:731–744.
Jeannesson-Thivisol E, Feillet F, Chery C et al. Genotype–phenotype associations in French patients with phenylketonuria and importance of genotype for full assessment of tetrahydrobiopterin responsiveness. Orphanet J Rare Dis 2015;10:158.
Guldberg P, Rey F, Zschocke J et al. A European multicenter study of phenylalanine hydroxylase deficiency: classification of 105 mutations and a general system for genotype-based prediction of metabolic phenotype. Am J Hum Genet 1998;63:71–79.
Zhu T, Ye J, Han L et al. Variations in genotype–phenotype correlations in phenylalanine hydroxylase deficiency in Chinese Han population. Gene 2013;529:80–87.
Santos LL, Fonseca CG, Starling AL et al. Variations in genotype–phenotype correlations in phenylketonuria patients. Genet Mol Res 2010;9:1–8.
The R Project for Statistical Computing 2017; http://www.R-project.org/ (accessed June 12, 2018)
Venables WN, Ripley BD, eds. Modern Applied Statistics with S. 4th ed. New York: Springer; 2002.
Russell L. emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 2018; https://CRAN.R-project.org/package=emmeans (accessed June 12, 2018).
Robin X, Turck N, Hainard A et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77.
Ritchie MD, Holzinger ER, Li R et al. Methods of integrating data to uncover genotype–phenotype interactions. Nat Rev Genet 2015;16:85–97.
Desviat LR, Perez B, Garcia MJ et al. Relationship between mutation genotype and biochemical phenotype in a heterogeneous Spanish phenylketonuria population. Eur J Hum Genet 1997;5:196–202.
Kayaalp E, Treacy E, Waters PJ et al. Human phenylalanine hydroxylase mutations and hyperphenylalaninemia phenotypes: a metanalysis of genotype–phenotype correlations. Am J Hum Genet 1997;61:1309–1317.
Bercovich D, Elimelech A, Zlotogora J et al. Genotype–phenotype correlations analysis of mutations in the phenylalanine hydroxylase (PAH) gene. J Hum Genet 2008;53:407–418.
Zhu T, Ye J, Han L et al. The predictive value of genetic analyses in the diagnosis of tetrahydrobiopterin (BH4)-responsiveness in Chinese phenylalanine hydroxylase deficiency patients. Sci Rep 2017;7:6762.
Cerone R, Andria G, Giovannini M et al. Testing for tetrahydrobiopterin responsiveness in patients with hyperphenylalaninemia due to phenylalanine hydroxylase deficiency. Adv Ther 2013;30:212–228.
Svensson E, von Dobeln U, Eisensmith RC et al. Relation between genotype and phenotype in Swedish phenylketonuria and hyperphenylalaninemia patients. Eur J Pediatr 1993;152:132–139.
Dobrowolski SF, Pey AL, Koch R et al. Biochemical characterization of mutant phenylalanine hydroxylase enzymes and correlation with clinical presentation in hyperphenylalaninaemic patients. J Inherit Metab Dis 2009;32:10–21.
Dobrowolski SF, Heintz C, Miller T et al. Molecular genetics and impact of residual in vitro phenylalanine hydroxylase activity on tetrahydrobiopterin-responsiveness in Turkish PKU population. Mol Genet Metab 2011;10:116–121.
Gersting SW, Kemter KF, Staudigl M et al. Loss of function in phenylketonuria is caused by impaired molecular motions and conformational instability. Am J Hum Genet 2008;83:5–17.
Blau N, Martinez A, Hoffmann GF et al. DNAJC12 deficiency: a new strategy in the diagnosis of hyperphenylalaninemias. Mol Genet Metab 2018;123:1–5.
Zhang Z, Gao JJ, Feng Y, et al. Mutational spectrum of the phenylalanine hydroxylase gene in patients with phenylketonuria in the central region of China. Scand J Clin Lab Invest 2018;78:211–218.
Wevers RA, Blau N, Think big—think omics. J Inherit Metab Dis 2018;41:281–283.
ACKNOWLEDGEMENTS
This work is part of the RD-CONNECT initiative and was supported by the FP7-HEALTH-2012-INNOVATION-1 EU Grant No. 305444 (to N.B.) and funding from the Dietmar-Hopp Foundation (to G.F.H. and N.B.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
DISCLOSURE
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Garbade, S.F., Shen, N., Himmelreich, N. et al. Allelic phenotype values: a model for genotype-based phenotype prediction in phenylketonuria. Genet Med 21, 580–590 (2019). https://doi.org/10.1038/s41436-018-0081-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0081-x
Keywords
This article is cited by
-
Allelic phenotype prediction of phenylketonuria based on the machine learning method
Human Genomics (2023)
-
Phenotypic correlates of structural and functional protein impairments resultant from ALDH5A1 variants
Human Genetics (2023)
-
Genetic etiology and clinical challenges of phenylketonuria
Human Genomics (2022)
-
Characterization of phenylalanine hydroxylase gene variants and analysis of genotype–phenotype correlation in patients with phenylalanine hydroxylase deficiency from Fujian Province, Southeastern China
Molecular Biology Reports (2022)
-
Italian national consensus statement on management and pharmacological treatment of phenylketonuria
Orphanet Journal of Rare Diseases (2021)