Abstract
Purpose
To determine the expressions of SARS-CoV-2 receptor angiotensin-converting enzyme 2 (ACE2) and type II transmembrane serine protease (TMPRSS2) genes in human and mouse ocular cells and comparison to other tissue cells.
Methods
Human conjunctiva and primary pterygium tissues were collected from pterygium patients who underwent surgery. The expression of ACE2 and TMPRSS2 genes was determined in human primary conjunctival and pterygium cells, human ocular and other tissue cell lines, mesenchymal stem cells as well as mouse ocular and other tissues by reverse transcription-polymerase chain reaction (RT-PCR) and SYBR green PCR.
Results
RT-PCR analysis showed consistent expression by 2 ACE2 gene primers in 2 out of 3 human conjunctival cells and pterygium cell lines. Expression by 2 TMPRSS2 gene primers could only be found in 1 out of 3 pterygium cell lines, but not in any conjunctival cells. Compared with the lung A549 cells, similar expression was noted in conjunctival and pterygium cells. In addition, mouse cornea had comparable expression of Tmprss2 gene and lower but prominent Ace2 gene expression compared with the lung tissue.
Conclusion
Considering the necessity of both ACE2 and TMPRSS2 for SARS-CoV-2 infection, our results suggest that conjunctiva would be less likely to be infected by SARS-CoV-2, whereas pterygium possesses some possibility of SARS-CoV-2 infection. With high and consistent expression of Ace2 and Tmprss2 in cornea, cornea rather than conjunctiva has higher potential to be infected by SARS-CoV-2. Precaution is necessary to prevent possible SARS-CoV-2 infection through ocular surface in clinical practice.
Similar content being viewed by others
Introduction
An outbreak of unknown causes of pneumonia cases was first identified in Wuhan, China in December 2019, with clinical presentations similar to viral pneumonia [1]. Genomic analysis on the RNA samples extracted from the bronchoalveolar-lavage fluid of the pneumonia patients by Illumina sequencing and nanopore sequencing identified the viral contigs matched to the genome of lineage B of the genus betacoronavirus and with 85% identity to the genome of a bat severe acute respiratory syndrome (SARS)-like coronavirus (bat-SL-CoVZC45, MG772933.1) [2], indicating that this coronavirus disease (COVID-19) is caused by a novel SARS coronavirus (SARS-CoV)-resembled virus (SARS-CoV-2). Target cell infection by SARS-CoV is mediated by the spike protein of coronavirus and the host cellular receptor, angiotensin-converting enzyme 2 (ACE2) [3]. Similarly, a recent study suggests that the cellular entry by SARS-CoV-2 also depends on ACE2 as well as type II transmembrane serine protease (TMPRSS2) [4]. This implies that the identification of ACE2 and TMPRSS2 expression in human tissues can predict the potential infected cells and their respective influences in COVID-19 patients.
Our center previously reported that no SARS-CoV can be found in tear swab and conjunctival scraping by reverse transcription-polymerase chain reaction (RT-PCR) or isolated by viral culture [5], and the SARS patients also did not show any observable ocular manifestations [6]. In contrast, conjunctivitis was suggested as a pre-symptomatic sign before the onset of COVID-19, possibly by the exposure of infectious droplets without proper protection of goggles [7]. Critically, SARS-CoV-2 can be detected in tears and conjunctival secretions from a COVID-19 patient with conjunctivitis by RT-PCR [8]. However, whether SARS-CoV-2 could infect human conjunctival cells and cause conjunctivitis still remain elusive. Herein, this study aimed to determine the expressions of SARS-CoV-2 receptor ACE2 and serine protease TMPRSS2 genes in human and mouse ocular cells and tissues. Their expressions were also compared with the expressions in other tissue cells.
Materials and methods
Human primary conjunctival and pterygium cell culture
The study protocol was approved by the Ethics Committee for Human Research of the Chinese University of Hong Kong and Joint Shantou International Eye Center of Shantou University and the Chinese University of Hong Kong, which is in accordance with the tenets of the Declaration of Helsinki. Informed consent was obtained from all study subjects before inclusion in the study. Human primary conjunctival and pterygium cell lines have been established and characterized as previously described [9, 10]. Briefly, full-length primary pterygium and conjunctiva tissues, freshly collected from surgical removal, were first digested with 50 μg/ml dispase (Gibco®, Rockville, MD) and 100 mM D-sorbitol (Sigma-Aldrich) in Dulbecco’s Modified Eagle Medium (DMEM; Gibco®) at 37 °C for 1 h, and further dissociated into single cells by 0.05% trypsin at 37 °C for 5 min with the cell strainer (40 μm, NuncTM). The isolated cells were cultured in advanced DMEM medium supplemented with 5% fetal bovine serum (FBS; Gibco®) and 1x penicillin-streptomycin (Gibco®) in 5% CO2 incubator at 37°C. The medium was changed in every 2–3 days. The conjunctival cells and pterygium cells with passage 3 were used in this study.
Human cell lines and mouse tissues
Human ocular cell lines, including retinal pigment epithelial (RPE) cells ARPE-19 (catalogue number: CRL-2302, American Type Culture Collection (ATCC), Manassas, VA) [11, 12], lens epithelial cells B3 (catalogue number: CRL-11421, ATCC) [13], primary trabecular meshwork (TM) cells [14], and retinoblastoma cells Y79 (catalogue number: HTB-18, ATCC) [15], have been previously established. We also collected human mesenchymal stem cells (MSCs) from different sources, including bone marrow-derived MSCs (BM-MSCs; catalogue number: 7500; ScienceCell Research Laboratories, Carlsbad, CA) [16], adipose-derived stem cells (ASCs) [17], and periodontal ligament-derived stem cells (PDLSCs) [18, 19]. Other human tissue cell lines, such as human umbilical vein endothelial cells (HUVEC; catalogue number: CRL-1730, ATCC), lung epithelial carcinoma cells A549 (catalogue number: CCL-185, ATCC), liver hepatocellular carcinoma cells HepG2 (catalogue number: HB-8065, ATCC) and epidermal keratinocytes HaCaT (catalogue number: T0020001, AddexBio, San Diego, CA), were also investigated in this study.
The study protocol was approved by the Animal Experimentation Ethics Committee of Joint Shantou International Eye Center of Shantou University and the Chinese University of Hong Kong. All experiments were conducted according to the guidelines of the Association for Research in Vision and Ophthalmology for the Use of Animals in Ophthalmic and Vision Research. Eight-month old C57BL/6 mice (n = 3) were sacrificed, and the eyeballs were enucleated and dissected in RNA later Stabilization Solution (Invitrogen). Different ocular tissues, including cornea, iris, lens, retina, and optic nerve. In addition, other tissues were also collected, including lung, heart, kidney, liver, spleen, and brain. All tissues were immediately immersed into the TRIzol reagent (Invitrogen) after dissection and stored at −80oC before further experiments.
Gene expression analysis
The expression levels of human ACE2 and TMPRSS2 as well as mouse Ace2 and Tmprss2 genes were evaluated by RT-PCR and SYBR green PCR analyses. Briefly, total RNA was extracted from the cells or tissues and purified with the TRIzol reagent according to the manufacturer’s protocol (Invitrogen). total RNA (500 ng) was reverse transcribed by SuperScript® III reverse transcriptase (Invitrogen). The genes were amplified by with specific primers (Supplementary Table 1). The housekeeping β-actin gene human ACTB and mouse Actb was used for normalization. The relative expression levels (ΔCt) of different human cell lines and mouse tissues were compared with that of A549 cells and mouse lung tissue respectively.
Statistical analysis
Each experiment was repeated three times. The expression data were presented as mean ± standard deviation. Mean of the three repeated experiments was compared by one-way analysis of variance with post-hoc Tukey’s test for multiple testing correction. All statistical analyses were performed by the commercially available software (IBM SPSS Statistics 22; SPSS Inc., Chicago, IL). Significance was defined as p < 0.05.
Results
Expression of ACE2 and TMPRSS2 genes in human conjunctival and pterygium cells
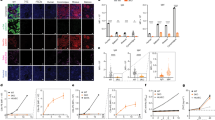
RT-PCR analysis showed that both ACE2 primers consistently amplified the ACE2 gene in 2 out of 3 conjunctival and pterygium cell lines (Fig. 1a). Consistent expression of ACE2 gene could be found in TM, HUVEC, HaCaT, PDLSC, A549, and HepG2 cells. HaCaT cells showed strongest expression of ACE2 among the tested cell lines, followed by HUVEC and HepG2 cells. Conjunctival cells moderately expressed ACE2, where pterygium cells showed weak expression. On the contrary, consistent expression of TMPRSS2 gene could only be found in 1 out of 3 pterygium cell lines, and no consistent expression of TMPRSS2 gene was observed in any conjunctival cells (Fig. 1b). In addition, consistent TMPRSS2 expression could be found in HUVEC, HaCaT, and A549 cells. Among the tested cell lines, HUVEC showed strongest expression of TMPRSS2 gene.
Full-length primary pterygium and conjunctiva tissues (each n = 3) were collected, dissociated and maintained in cell culture. Human ocular cell lines, including retinal pigment epithelial cells ARPE-19, lens epithelial cells B3, primary trabecular meshwork (TM) cells and retinoblastoma cells Y79, human primary mesenchymal stem cells (MSCs), including bone marrow-derived MSCs (BM-MSCs), adipose-derived stem cells (ASC), and periodontal ligament-derived stem cells (PDLSC), as well as other human tissue cell lines, including human umbilical vein endothelial cells (HUVEC), lung epithelial carcinoma cells A549, liver hepatocellular carcinoma cells HepG2 and keratinocytes HaCaT, were also culture and collected. Total RNA was extracted and reverse transcribed (RT). The expression of (a) ACE2 and (b) TMPRSS2 genes was determined by polymerase chain reaction (PCR) with 2 primers for each gene. Housekeeping β-actin gene ACTB was used for normalization.
SYBR green PCR analysis confirmed that HaCaT cells had higher expression of ACE2 gene with the ΔCt of 9.01 ± 0.10 and 7.44 ± 0.04 for primers 1 and 2, respectively as compared with other cell lines (Fig. 2a). Compared with the lung epithelial carcinoma cells A549, HaCaT cells (primer 1: 801.6-fold, p < 0.001; primer 2: 187.2-fold, p < 0.001) and ASCs (primer 1: 6.4-fold, p < 0.001; primer 2: 7.5-fold; p = 0.034) consistently showed significant higher expression for the 2 ACE2 primers, whereas human TM cells consistently showed significant lower expression (primer 1: 3.8-fold, p = 0.027; primer 2: 6.2-fold, p = 0.048). In contrast, 2 conjunctival cell lines, ARPE-19, HUVEC, and HepG2 cells only showed differential expression in the ACE2 gene amplification by primer 1 as compared with the A549 cells (p < 0.05). There was no consistent significant differential expression of TMPRSS2 gene as compared with the A549 cells (Fig. 2b). Pterygium sample 3 and ASCs showed significant higher TMPRSS2 expression in primer 1, whereas BM-MSCs, B3, and Y79 cells showed significant lower expression in primer 2. The conjunctival cells showed similar expression of TMPRSS2 gene as compared with the A549 cells.
Full-length primary pterygium and conjunctiva tissues (each n = 3) were collected, dissociated and maintained in cell culture. Human ocular cell lines, including retinal pigment epithelial cells ARPE-19, lens epithelial cells B3, primary trabecular meshwork (TM) cells and retinoblastoma cells Y79, human primary mesenchymal stem cells (MSCs), including bone marrow-derived MSCs (BM-MSCs), adipose-derived stem cells (ASC), and periodontal ligament-derived stem cells (PDLSC), as well as other human tissue cell lines, including human umbilical vein endothelial cells (HUVEC), lung epithelial carcinoma cells A549, liver hepatocellular carcinoma cells HepG2 and keratinocytes HaCaT, were also culture and collected. Total RNA was extracted and reverse transcribed. The expression of (a) ACE2 and (b) TMPRSS2 genes was determined by SYBR green polymerase chain reaction (PCR) with 2 primers for each gene. Housekeeping β-actin gene ACTB was used for normalization. Relative expression levels (ΔCt) of different mouse tissues were compared with that of A549 cells. *p < 0.05; **p < 0.01; ***p < 0.001.
Expression of Ace2 and Tmprss2 genes in mouse ocular tissues
All collected mouse tissues could express the Ace2 gene by the 2 primers in the RT-PCR analysis (Fig. 3a). Stronger expression of Ace2 gene could be found in cornea and liver among the tested tissues, whereas lung, heart, kidney, and brain showed moderate expression. In contrast, expression of Tmprss2 gene could only consistently be found in mouse cornea, lung, liver, kidney, and brain tissues (Fig. 3b). Cornea and liver had stronger expression of Tmprss2 gene among the collected tissues, whereas lung and kidney showed moderate expression.
Eight-month-old C57BL/6 mice (n = 3) were sacrificed, and different ocular tissues, including cornea, iris, lens, retina and optic nerve, as well as other tissues, including lung, heart, kidney, liver, spleen, and brain, were collected. Total RNA was extract and reverse transcribed (RT). The expression of (a) Ace2 and (b) Tmprss2 genes was determined by polymerase chain reaction (PCR) with 2 primers for each gene. Housekeeping β-actin gene Actb was used for normalization.
SYBR green PCR analysis confirmed the expression of Ace2 gene in all collected mouse tissues (Fig. 4a), with stronger expression in lung (primer 1 ΔCt: 7.76 ± 0.95 and primer 2 ΔCt: 8.33 ± 0.18) and kidney (primer 1 ΔCt: 5.26 ± 0.24 and primer 2 ΔCt: 4.79 ± 0.25). Compared with the lung tissue, only kidney tissue consistently showed significant higher expression for the 2 Ace2 primers by 5.7 and 11.6-fold, respectively (p < 0.01). Other tissues, except heart, showed significantly lower Ace2 gene expression than the lung (p < 0.01). Cornea tissue had 8.7 (p < 0.001) and 4.3-fold (p = 0.001) lower expression for the Ace2 primer 1 and primer 2 respectively as compared with the lung tissue. For the Tmprss2 gene, the expression could be found in all collected mouse tissues (Fig. 4b). Strongest expression was observed in kidney tissue with the ΔCt of 3.55 ± 0.11 and 3.37 ± 0.10 for primer 1 and 2, respectively, followed by the lung, cornea, and liver. Coherent to the Ace2 gene, only kidney tissue consistently showed significant higher expression for the 2 Tmprss2 primers by 9.6 and 9.7-fold, respectively (p < 0.01) as compared with the lung tissue. In contrast, iris, lens, retina, optic nerve, heart, spleen, and brain showed significant lower Tmprss2 gene expression than the lung tissue. The expressions of Tmprss2 gene in cornea and liver were comparable to those in the lung tissue.
Eight-month-old C57BL/6 mice (n = 3) were scarified, and different ocular tissues, including cornea, iris, lens, retina, and optic nerve, as well as other tissues, including lung, heart, kidney, liver, spleen, and brain, were collected. Total RNA was extracted and reverse transcribed. The expression of (a) Ace2 and (b) Tmprss2 genes was determined by SYBR green polymerase chain reaction (PCR) with 2 primers for each gene. Housekeeping β-actin gene Actb was used for normalization. Relative expression levels (ΔCt) of different mouse tissues were compared with that of mouse lung tissue. *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
Results from this study showed that: (1) consistent expression of ACE2 and TMPRSS2 genes could only be observed in the conjunctival cells and pterygium cells from some pterygium patients; (2) mouse cornea had comparable expression of Tmprss2 gene and lower but prominent Ace2 gene expression than the lung tissue; (3) high expression of Ace2 and Tmprss2 genes could be found in mouse cornea. Collectively, our results indicate that, with reference to the Ace2 and Tmprss2 expression in the lung, cornea rather than conjunctiva could potentially be infected by SARS-CoV-2.
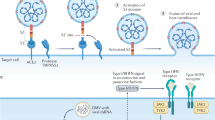
COVID-19 pandemic is a global issue. More than 200 countries/territories have been affected, with more than 1 million confirmed cases. SARS-CoV-2 has been discovered as the pathogenic cause of COVID-19 [2]. It is believed that person-to-person transmission of COVID-19 is mainly due to the infection by SARS-CoV-2 through respiratory droplets and close contact [20]. The infection and cellular entry of SARS-CoV-2 are mediated by the spike glycoprotein of coronavirus and the host cellular SARS-CoV receptor ACE2 [4, 21]. In addition to ACE2, TMPRSS2 is required to promote SARS-CoV entry by ACE2 cleavage to promote viral uptake and SARS-CoV spike protein cleavage to activates the spike protein for membrane fusion [22]. Therefore, understanding the coexpression of ACE2 and TMPRSS2 can predict the possibility of SARS-CoV-2 infection in human system.
In this study, we, for the first time, demonstrated ACE2 gene expression in some but not all primary human conjunctival and pterygium cells (Fig. 1a). Similarly, consistent expression of TMPRSS2 gene could only be found in 1 out of 3 investigated pterygium samples, but not in any conjunctival cells (Fig. 1b). Considering the necessity of both ACE2 and TMPRSS2 for SARS-CoV-2 infection, our results suggest that conjunctiva would be less likely to be infected by SARS-CoV-2, whereas pterygium possesses some possibility of SARS-CoV-2 infection. On the contrary, consistent expression of ACE2 and TMPRSS2 genes can be found in endothelial cells HUVEC, epidermal keratinocytes HaCaT, and lung epithelial carcinoma cells A549, indicating that lung, skin, and blood capillaries are the potential sites with SARS-CoV-2 infection. To expand our understanding of the possible tissues influenced by SARS-CoV-2 infection, we determined the expression of Ace2 and Tmprss2 genes in mouse tissues (Fig. 3). We report the expression of Ace2 and Tmprss2 genes in mouse cornea tissue, but not in iris, lens, retina, and optic nerve. Our results indicated that cornea could be a susceptible site for SARS-CoV-2 infection as their expressions in cornea are comparable to that in lung tissue (Fig. 4). Apart from cornea, we found expression of Ace2 and Tmprss2 genes in lung, liver, kidney, and brain. A recent RNA sequencing analysis on 13 human tissues reports the expression of ACE2 mainly in lung AT2, liver cholangiocyte, colon colonocytes, oesophagus keratinocytes, ileum enterocytes, rectum enterocytes, stomach epithelial cells, and kidney proximal tubules, and ACE2 expression was also observed in pericytes and fibroblasts from the eye tissue [23]. ACE2 expression can also be noted in the epithelial cells of oral mucosa, vascular endothelial cells, type II pneumocytes, kidney, heart, and the central nervous system [24,25,26,27,28]. On the other hand, TMPRSS2 expression has been reported in primary human airway cells, type II pneumocytes and cancers [29, 30].
ACE2 is a negative regulator of the renin-angiotensin-aldosterone system, controlling the blood pressure, body fluid homeostasis as well as the inflammatory and fibrotic effects [31]. In the eye, ACE2 expression was found in ciliary body, retina, and photoreceptors [32, 33]. Activation of endogenous Ace2 by diminazene aceturate facilitates the drainage of aqueous humor and decreases intraocular pressure in a rat model of glaucoma [34]. Overexpression of ACE2 can also ameliorate the inflammatory response in experimental autoimmune uveitis model and amyloid β-treated human RPE [35, 36]. Although ACE2 is critical for SARS-CoV infection, SARS-CoV infection and spike protein of SARS-CoV would reduce ACE2 expression, and injection of SARS-CoV spike protein worsens acute lung failure in mice with the enhanced renin-angiotensin pathway [37]. Considering the correlation of reduced ACE2 expression after SARS-CoV infection and the anti-inflammatory role of ACE2, it could explain the possible occurrence of conjunctivitis in patients with SARS-CoV-2 infection [7, 8].
TMPRSS2, belonging to the TMPRSS2 family, cleaves the coronavirus spike protein for coronavirus infections, which lack of Tmprss2 reduces the severity of lung pathology after infection by SARS-CoV and Middle East Respiratory Syndrome-CoV [38]. In addition to the coronavirus spike protein, TMPRSS2 is also the major protease for the cleavage of the influenza virus surface glycoprotein hemagglutinin, which is prerequisite for membrane fusion and virus infectivity [29]. Apart from TMPRSS2, membrane alanyl aminopeptidase (ANPEP), dipeptidyl peptidase 4, and glutamyl aminopeptidase have been suggested as the candidate coreceptors for ACE2 [23]. The roles and mechanisms of these coreceptor peptidases in SARS-CoV-2 infection as well as the possible therapeutic treatments for COVID-19 warrant further investigations.
In summary, this study revealed the expression of SARS-CoV-2 receptor ACE2 and serine protease TMPRSS2 genes in human conjunctival and pterygium cells as well as mouse cornea tissue. Considering the high expression of Ace2 and Tmprss2 in cornea, precaution practices are necessary to prevent possible SARS-CoV-2 infection through ocular surface.
Summary
What was known before
ACE2 is a negative regulator of the renin-angiotensin-aldosterone system, controlling the blood pressure, body fluid homeostasis, inflammation, and fibrosis.
In the eye, ACE2 expression was found in ciliary body, retina, and photoreceptors.
What this study adds
Consistent expression of ACE2 and TMPRSS2 genes was only observed in the conjunctival cells and pterygium cells from some pterygium patients.
Mouse cornea had comparable expression of Tmprss2 gene and lower but prominent Ace2 gene expression than the lung tissue.
References
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506.
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33.
Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–4.
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80. e8.
Chan WM, Yuen KS, Fan DS, Lam DS, Chan PK, Sung JJ. Tears and conjunctival scrapings for coronavirus in patients with SARS. Br J Ophthalmol. 2004;88:968–9.
Yuen KS, Chan WM, Fan DS, Chong KK, Sung JJ, Lam DS. Ocular screening in severe acute respiratory syndrome. Am J Ophthalmol. 2004;137:773–4.
Lu CW, Liu XF, Jia ZF. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395:e39.
Xia J, Tong J, Liu M, Shen Y, Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J Med Virol. 2020 https://doi.org/10.1002/jmv.25725. [Epub ahead of print]. PubMed PMID: 32100876.
Yang Q, Jhanji V, Tan SQ, Chan KP, Cao D, Chu WK, et al. Continuous exposure of nicotine and cotinine retards human primary pterygium cell proliferation and migration. J Cell Biochem. 2019;120:4203–13.
Cao D, Ng TK, Yip YWY, Young AL, Pang CP, Chu WK, et al. p53 inhibition by MDM2 in human pterygium. Exp Eye Res. 2018;175:142–7.
Zhang XY, Ng TK, Brelén ME, Chan KP, Wu D, Yung JSY, et al. Disruption of retinal pigment epithelial cell properties under the exposure of cotinine. Sci Rep. 2017;7:3139.
Ng TK, Liang XY, Lu F, Liu DT, Yam GH, Ma L, et al. Protective effects of an HTRA1 insertion-deletion variant against age-related macular degeneration in the Chinese populations. Lab Invest. 2017;97:43–52.
Chen JH, Huang C, Zhang B, Yin S, Liang J, Xu C, et al. Mutations of RagA GTPase in mTORC1 pathway are associated with autosomal dominant cataracts. PLoS Genet. 2016;12:e1006090.
Wu Z, Huang C, Xu C, Xie L, Liang JJ, Liu L, et al. Caveolin-1 regulates human trabecular meshwork cell adhesion, endocytosis, and autophagy. J Cell Biochem. 2019;120:13382–91.
Chu WK, Law KS, Chan SO, Yam JC, Chen LJ, Zhang H, et al. Antagonists of growth hormone-releasing hormone receptor induce apoptosis specifically in retinoblastoma cells. Proc Natl Acad Sci USA. 2016;113:14396–401.
Ng TK, Carballosa CM, Pelaez D, Wong HK, Choy KW, Pang CP, et al. Nicotine alters MicroRNA expression and hinders human adult stem cell regenerative potential. Stem Cells Dev. 2013;22:781–90.
Huang Y, Ng TK, Chen CB, Huang B, Liang J, Pang CP, et al. Notch signaling activation enhances human adipose-derived stem cell retinal differentiation. Stem Cells Int. 2018;2018:9201374.
Cen LP, Ng TK, Liang JJ, Zhuang X, Yao X, Yam GH, et al. Human periodontal ligament-derived stem cells promote retinal ganglion cell survival and axon regeneration after optic nerve injury. Stem Cells. 2018;36:844–55.
Ng TK, Yung JS, Choy KW, Cao D, Leung CK, Cheung HS, et al. Transdifferentiation of periodontal ligament-derived stem cells into retinal ganglion-like cells and its microRNA signature. Sci Rep. 2015;5:16429.
Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–23.
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–8.
Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88:1293–307.
Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526:135–40.
Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:8.
Baig AM, Khaleeq A, Ali U, Syeda H. Evidence of the COVID-19 virus targeting the cns: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci. 2020;11:995–8.
Song J, Hu B, Qu H, Wang L, Huang X, Li M, et al. Upregulation of angiotensin converting enzyme 2 by shear stress reduced inflammation and proliferation in vascular endothelial cells. Biochem Biophys Res Commun. 2020;525:812–8.
Oudit GY, Kassiri Z, Jiang C, Liu PP, Poutanen SM, Penninger JM, et al. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur J Clin Invest. 2009;39:618–25.
Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–7.
Limburg H, Harbig A, Bestle D, Stein DA, Moulton HM, Jaeger J, et al. TMPRSS2 Is the major activating protease of influenza A virus in primary human airway cells and influenza B virus in human type II pneumocytes. J Virol. 2019;93:e00649–19.
Chen Z, Song X, Li Q, Xie L, Guo T, Su T, et al. Androgen receptor-activated enhancers simultaneously regulate oncogene TMPRSS2 and lncRNA PRCAT38 in prostate cancer. Cells. 2019;8:E864.
Mirabito Colafella KM, Bovée DM, Danser AHJ. The renin-angiotensin-aldosterone system and its therapeutic targets. Exp Eye Res. 2019;186:107680.
Luhtala S, Vaajanen A, Oksala O, Valjakka J, Vapaatalo H. Activities of angiotensin-converting enzymes ACE1 and ACE2 and inhibition by bioactive peptides in porcine ocular tissues. J Ocul Pharm Ther. 2009;25:23–28.
Tikellis C, Johnston CI, Forbes JM, Burns WC, Thomas MC, Lew RA, et al. Identification of angiotensin converting enzyme 2 in the rodent retina. Curr Eye Res. 2004;29:419–27.
Foureaux G, Nogueira JC, Nogueira BS, Fulgêncio GO, Menezes GB, Fernandes SO, et al. Antiglaucomatous effects of the activation of intrinsic angiotensin-converting enzyme 2. Invest Ophthalmol Vis Sci. 2013;54:4296–306.
Qiu Y, Tao L, Zheng S, Lin R, Fu X, Chen Z, et al. AAV8-mediated angiotensin-converting enzyme 2 gene delivery prevents experimental autoimmune uveitis by regulating MAPK, NF-κB and STAT3 pathways. Sci Rep. 2016;6:31912.
Fu X, Lin R, Qiu Y, Yu P, Lei B. Overexpression of angiotensin-converting enzyme 2 ameliorates amyloid β-induced inflammatory response in human primary retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2017;58:3018–28.
Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–9.
Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93:e01815–18.
Acknowledgements
We would like to express our deepest gratitude to all study participants. This study was supported by an internal grant from the Joint Shantou International Eye Center of Shantou University and the Chinese University of Hong Kong (20–015 to TKN), and the Grant for Key Disciplinary Project of Clinical Medicine under the Guangdong High-level University Development Program (002–18119101), China.
Author information
Authors and Affiliations
Contributions
TKN conception and design. TKN financial support. DM, VJ, YH, LPC, and TKN provision of study materials. CBC, CX, and JJL collection and/or assembly of data. CBC and TKN data analysis and interpretation. VJ, XLY, and TKN manuscript writing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ma, D., Chen, CB., Jhanji, V. et al. Expression of SARS-CoV-2 receptor ACE2 and TMPRSS2 in human primary conjunctival and pterygium cell lines and in mouse cornea. Eye 34, 1212–1219 (2020). https://doi.org/10.1038/s41433-020-0939-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41433-020-0939-4
This article is cited by
-
Effects of particulate matter exposure on the expression of the SARS-CoV-2 ACE2 receptor in ocular surface tissues and cells
Environmental Science and Pollution Research (2024)
-
High-throughput screening of genetic and cellular drivers of syncytium formation induced by the spike protein of SARS-CoV-2
Nature Biomedical Engineering (2023)
-
Two years of SARS-CoV-2 infection (2019–2021): structural biology, vaccination, and current global situation
The Egyptian Journal of Internal Medicine (2022)
-
Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture
Communications Biology (2022)
-
SARS-COV-2 viral load in tears of patients with COVID-19 in the early symptomatic stages: comparison of two different tear sampling methods
International Ophthalmology (2022)