Abstract
Alkaptonuria (AKU) is a rare metabolic disorder caused by a deficient enzyme in the tyrosine degradation pathway, homogentisate 1,2-dioxygenase (HGD). In 172 AKU patients from 39 countries, we identified 28 novel variants of the HGD gene, which include three larger genomic deletions within this gene discovered via self-designed multiplex ligation-dependent probe amplification (MLPA) probes. In addition, using a reporter minigene assay, we provide evidence that three of eight tested variants potentially affecting splicing cause exon skipping or cryptic splice-site activation. Extensive bioinformatics analysis of novel missense variants, and of the entire HGD monomer, confirmed mCSM as an effective computational tool for evaluating possible enzyme inactivation mechanisms. For the first time for AKU, a genotype–phenotype correlation study was performed for the three most frequent HGD variants identified in the Suitability Of Nitisinone in Alkaptonuria 2 (SONIA2) study. We found a small but statistically significant difference in urinary homogentisic acid (HGA) excretion, corrected for dietary protein intake, between variants leading to 1% or >30% residual HGD activity. There was, interestingly, no difference in serum levels or absolute urinary excretion of HGA, or clinical symptoms, indicating that protein intake is more important than differences in HGD variants for the amounts of HGA that accumulate in the body of AKU patients.
Similar content being viewed by others
Introduction
Alkaptonuria (AKU) [OMIM 203500] was the first genetic disease identified [1], which is characterized by deficiency of homogentisate 1,2-dioxygenase (HGD) (EC 1.13.11.5), an enzyme involved in the metabolism of tyrosine [2]. AKU is very rare in most ethnic groups (1:1,000,000–250,000), but in some countries it exhibits increased prevalence [3,4,5,6].
The metabolic block in AKU causes accumulation of homogentisic acid (HGA). Large amounts of HGA are excreted in the urine, resulting in characteristic darkening upon standing [7]. Excess HGA that is not renally excreted accumulates in the body where it polymerizes, forming a dark brown ochronotic pigment that is deposited in connective tissue, mainly in the skin, sclera, spine, and large-joint cartilage, as well as in heart valves, where it causes aortic stenosis. AKU patients suffer from early onset severe arthropathy, usually starting in their early 30s. Manifestation of the disease varies between individual patients and increases with age as a result of ongoing HGA accumulation. No specific cure exists for this disorder; painkillers and joint replacement surgery in advanced stages are the only palliative treatments [8].
AKU patients carry homozygous or compound heterozygous variants of the HGD gene [4, 9] (3q13.33) a single-copy gene composed of 14 exons [10,11,12]. To date, DNA sequencing has been performed in ~400 AKU patients, leading to the identification of 175 different HGD variants, of which 142 are most likely disease-causing, variants that affect function [13]. All variants identified in AKU patients worldwide are summarized in the HGD mutation database (http://hgddatabase.cvtisr.sk/) [14].
The HGD protein protomer is composed of 445 amino acids (NP_000178.2) and is expressed in the prostate, small intestine, colon, kidney, and liver [10], as well as in osteoarticular compartment cells (chondrocytes, synoviocytes, and osteoblasts) [15]. The experimental crystal structure of the HGD protein has been solved (PDB code 1EY2 and 1EYB), revealing that the active form of the enzyme is organized as a highly complex and dynamic hexamer comprising two disk-like trimers [16]. An intricate network of noncovalent interactions is required to maintain the spatial structure of the protomer, the trimer and, finally, the hexamer. This delicate structure has a low tolerance to mutation and can be easily disrupted mainly by missense variants (representing ~68% of all known AKU variants) [13, 17] compromising enzyme function. Recently, we showed that the missense variants are predicted to affect the activity of the enzyme by three molecular mechanisms: decrease of stability of individual protomers, disruption of protomer–protomer interactions or modification of residues in the active site region [13].
The currently ongoing DevelopAKUre project consist of three studies aimed to test nitisinone for AKU. The first one was Suitability Of Nitisinone In Alkaptonuria 1 (SONIA1) study that confirmed nitisinone decreases urine HGA in a dose-dependent, as well as a concentration-dependent manner [18, 19]. The SONIA2 long-term study, which is due to finish in 2019, aims to assess the efficacy of nitisinone treatment on clinical outcome, biochemical markers and safety. The Subclinical Ochronotic Features In Alkaptonuria (SOFIA) study evaluates at what age ochronosis starts and whether it presents before the onset of clinical symptoms of AKU, such as joint pain. In addition, we have established a novel ApreciseKUre database that facilitates collection and analysis of clinical and biochemical patient data shared among registered researchers [20].
In the present report, we analyzed samples of 172 AKU patients, including those from the SONIA2 and SOFIA studies. For the first time in AKU, we performed HGD gene sequencing, as well as multiplex ligation-dependent probe amplification (MLPA) analysis in order to uncover possible larger genomic deletions, and we tested several intronic and exonic variants for their possible effect on splicing. We used SONIA2 baseline clinical data to test for possible genotype–phenotype correlations.
Subjects and methods
AKU patients
DNA samples from 172 AKU patients (105 males, 67 females) were collected in our laboratory and analyzed by genomic sequencing and MLPA analysis. Informed consent was obtained from all patients listed in S1 Table with patient codes P1-172. DNAs of the patients P1-24 were sent for mutation analysis to our lab directly, patients P25-P33 were enrolled in the SOFIA study in Liverpool, and patient P34-172 were enrolled in the SONIA2 study in one of three clinical trial centers: Liverpool (P34-74), Piestany (P75-139), and Paris (P140-172). AKU diagnosis was established based on documented elevated HGA in urine and/or the bluish-black pigmentation in connective tissue (ochronosis). All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000.
HGD variants and MLPA analysis
All 14 exons of the HGD gene were screened for variants by DNA sequencing per patient as described previously [14]. Ten patients, in whom no variant was identified using sequencing, were analyzed by MLPA analysis (MRC Holland) using our self-designed, HGD-gene, synthetic probes (Zatkova et al., Unpublished data). Novel variants were reported according to the Human Genome Variation Society (HGVS) nomenclature additions [21] and their description is based on coding DNA Reference Sequence NM_000187.3 (genomic reference sequence NG_011957.1). Exons were numbered as by Granadino and colleagues [12]. Variant nomenclature was verified using MUTALYSER Name Checker (https://mutalyzer.nl/). All novel variants as well as all individual patients were deposited in the HGD gene mutation database [14] (http://hgddatabase.cvtisr.sk/), using a specific DB_ID for each variant (Table 1) and unique family/allele code for each patient (S1 Table). In order to facilitate variant recognition, the tables also include the short names used by the AKU scientific community.
Evaluation of the effect of missense variants on HGD protein function
The possible effect of the novel missense variants on HGD protein function was tested as described previously [13]. Details can be found in Supplementary Methods.
In silico predictions of variant-induced alterations in exonic and intronic splicing regulatory sequences
Potential impact of HGD variants on RNA splicing was based on the density calculation of auxiliary splicing motifs, using tools described in Supplementary Methods.
Plasmid constructs for a splicing reporter minigene assay
We used the U2AF1 splicing reporter, which has been previously described [22]. The pU2AF1-HGD E6, pU2AF1-HGD E9, and pU2AF1-HGD E10 three-exon minigene constructs (Fig. 1a) contained genomic segments comprising HGD exon 6, exon 9, and exon 10, respectively, and part of the 5′ and 3′ flanking intronic sequences. The pU2AF1-HGD E14 (Fig. 1b) is a two-exon minigene that contains 303nt of the last exon 14 and part of the 5′ flanking intronic sequence. Details can be found in Supplementary Methods.
Analysis of HGD variants with splicing reporter minigene assays in HepG2 cells. Schematic representation of (a) pU2AF1-HGD E6, pU2AF1-HGD E9, and pU2AF1-HGD E10 minigenes and (b) pU2AF1-HGD E14 minigene. XhoI/XbaI segments of HGD, containing exon 6, 9, or exon 10 (black box) and XhoI/ApaI segment of exon14 (textured box), were cloned between U2AF1 exons 2 and 4 (white boxes) or between U2AF1 exons 2 and MCS of pcDNA3.1vector, respectively. RT-PCR primers to amplify reporter transcripts are denoted by arrows. (c) Splicing pattern of wild-type and mutated HGD minigenes from panels a and b. Variants are at the top of panel; RNA products are shown schematically to the right. EI exon inclusion. c.650-17G>A shows almost a complete exon skipping, while c.650-13T>G leads to a mixture of skipped and correctly spliced products and activation of a cryptic 3′ ss-176 (CR3′ss-176). In case of c.650-56G>A variant EI is decreased to 73% and use of a cryptic 3′ ss-176 (CR3′ss-176) is more evident. c.649+39T>G creates de novo donor splice site at position +1 relative to 5′ss consensus sequence that is stronger than the authentic counterparts and causes inclusion of part of the intron 9 into the transcript
Cell cultures and transfections
HEK293 (human embryonic kidney) and HepG2 (liver hepatocellular carcinoma) cells were grown under standard conditions, transfected with individual minigene construct and harvested 24 h later. Total RNA was extracted, reverse-transcribed and complementary DNA samples were amplified using PL3 and PL4 [23], or PL5 (CAG GTG CTC TCG GTT GCA) and PL4 primers. Spliced products were gel separated and sequenced to determine their identity. Details can be found in Supplementary Methods.
Analysis of general HGD protein structure and its conservation
The Homo sapiens HGD (HGDHs) protein sequence (code Q93099) was used to search for homologous proteins, with which subsequently multiple alignments were performed and for each position, Shannon Entropy values were computed by Bio3D [24] in order to assess conservation across evolution. For the sake of clarity, the complementary Shannon entropy values were reported as H.10 and normalized to 1 (i.e., a value of 0 corresponds to maximum conservation and 1 to maximum variability). We also considered population-based variability using the missense tolerance ratio (MTR) scoring system [25]. Finally, the secondary structure was assigned to HGD amino acids using define secondary structure of protein (DSSP) [26].
Structural reconstruction of HGD monomer
The structures of holo-HGDHs (PDB ID: 1EY2) and its homologous Pseudomonas putida HGD (HGDPp) (PDB ID: 4AQ2) were retrieved from the Protein Data Bank (PDB) [27]. The missing segments (loop) in the human protein structure (residues 348–355) were reconstructed by homology modeling using the HGDPp structure. Details can be found in Supplementary Methods. The completed reconstructed monomer structure served as a starting point for the reconstruction of the entire HGDHs oligomeric protein, as well as for all predictions.
Molecular dynamics of the HGD hexamer and the analysis of inter-trimeric and intra-trimeric interactions
A molecular dynamics simulation of the hexameric structure of HGD was performed using GROMACS 5.0.2 [28]. In PyMOL v1.7.2.1 [29] we selected and further investigated for inter-protomer H-bonding by the gmx H-bond all the residues within 5 Å of chain A. Other noncovalent interactions at the protomer–protomer interface, including van der Waals and pi stacking, were evaluated as a whole by use of contact map, averaged along the simulation trajectory and with a cut-off of 1.5 nm to include interactions up to the ionic pairing. In order to investigate structural effects of the three most frequent variants, M368V, G161R, and A122V, molecular dynamics simulations were carried out on three-dimensional structures of native and mutated HGD. Details can be found in Supplementary Methods.
Functional analysis of variants G161R, M368V, and A122V
Recombinant human HGD, with an N-terminal deca-His tag followed by a TEV protease cleavage site, was cloned into a pET21a vector and transformed into E. coli BL21(DE3) pLysS. Cells were grown, and after the cell lysis and recombinant protein extraction and purification the HGD enzyme activity was measured according to previously published protocols [30]. SEC-MALS analysis was performed to determine the experimental molecular weight of the recombinant HGD complexes as previously described [31,32,33,34]. Details can be found in Supplementary Methods.
Genotype–phenotype correlation studies
Genotype–phenotype correlation studies were carried out in 33 of the 139 patients participating in the SONIA2 study. Serum and urine samples were obtained from each patient at the baseline visit in the three clinical trial centers and serum and urine HGA (s-HGA, u-HGA) quantitation analyses were performed as previously described [18, 35, 36]. The urinary excretion of HGA during 24 h (u-HGA24) was determined and was also corrected for urinary excretion of urea, an approximate measure of protein intake, in order to compensate for differences in protein (tyrosine) dietary intake (u-HGA/u-urea ratio). We compared s-HGA, u-HGA24, and u-HGA/u-urea for patients who are homozygous for either the G161R variant (Group A, 16 patients, a variant with low residual HGD activity: 1% of wild type) or the A122V or M368V variants (Group B, 17 patients, variants with relatively high residual enzyme activity: >30% of wild type). In addition, we checked for possible differences between Groups A and B in eye pigmentation, bone density of the hip, Cobbs angle for scoliosis, and peak aortic velocity (severity of aortic stenosis), all measured at baseline in the SONIA2 study. Details can be found in Supplementary methods. Comparisons between Groups A and B were made using a two-tailed, two-sample, and equal variance t-test.
Results and discussion
28 novel variants identified
We identified 73 different variants across the 172 AKU patients, 28 variants were novel (Table 1). Thus, the total number of HGD gene variants identified in AKU to date, and listed in the HGD mutation database, is 203. Fifteen of the 28 novel variants led to an amino acid substitution and were predicted as likely to affect function by PolyPhen-2 and/or SNAP (Table 1). The most prevalent was the G161R missense variant in exon 8, found in 68 AKU chromosomes, mainly in patients from Slovakia, followed by a missense variant A122V in exon 6 (27 AKU chromosomes, mainly from Jordan), and M368V in exon 13 (24 AKU chromosomes), the most frequent European variant. Fifty-three of the identified variants were present in only one or two AKU chromosomes, confirming a high genetic heterogeneity of AKU. In one SONIA2 patient from Romania (P62), three variants were present, two copies of c.16-1G>A (ivs1-1G>A), and one copy of D153fs (S1 Table).
Despite lacking any typical exonic variant, one patient from Mali (P171) was homozygous for two novel intronic variants: c.343-11 G>A (ivs5-11G>A) and c.650-13 T>G (ivs9-13T>G) (S1 Table). Elsewhere, four additional intronic variants have been reported previously, in which an effect on splicing was proposed as the likely mutation mechanism: c.650-85 A>G (AKU_DB_183) [13], c.650-17 G>A (AKU_DB_10) [37], c.650-56G>A (AKU_DB_10, AKU_DB_11, P21) [37], and c.649+39T>G (AKU_DB_131, AKU_DB_219) [38]. The intronic variants c.343-11G>A, c.650-13T>G, and c.650-17G>A are located in the Y-rich polypyrimidine tract regulatory sequence upstream of the 3′ splice site (3′ ss), and were predicted to reduce the strength of the 3′ ss (data not shown). The c.650-17G>A substitution creates a potential 3′ AG site, located 11 nucleotides from the predicted branch point (ivs9-27 (c.650-27), SVM BP score 0.65), thus this AG-creating variant is located within the critical distance from the predicted branch point required for efficient recognition of 3′ ss and may be sufficient to suppress utilization of wild-type 3′ ss [39]. For the variant c.650-85A>G, the HSF predicted activation of a cryptic splice site, which would lead to insertion of 28 amino acids into the HGD protein sequence at the homohexamer interface, disrupting formation of the complex [13]. In addition, we identified silent variants c.372C>T (exon 6) and c.1191A>C (exon 14), both suspected by HSF analysis to abolish or reduce the strength of the predicted ESEs (data not shown).
Since patient tissue expressing HGD protein was not available (e.g., cartilage, liver, kidney), we used minigene constructs to test the possible effect of all eight variants on mRNA splicing. Minigene experiments revealed that c.650-13T>G and c.650-17G>A led to a splicing defect in both HepG2 and HEK293 cells (Fig. 1a, c), supporting classification as variants that affect HGD function. Although the close proximity of these variants, c.650-17G>A produced almost complete exon 10 skipping, while c.650-13T>G lead to a mixture of skipped and correctly spliced products and activation of a cryptic 3′ ss-176. Variant c.650-56G>A reduced exon skipping but activated cryptic 3′ ss-176 to greater extent (Fig. 1c). Despite the formation of a 3′ AG by c.650-17G>A substitution, this AG was not used. Single base change c.649+39T>G-induced aberrant splicing by using de novo donor splice site created by this variant at position +1 relative to 5′ss consensus sequence that was stronger than the authentic counterparts (MAXENT: 8.24 vs. 4.94). This effect was evident in HepG2 cells (Fig. 1c).
Variants c.343-11G>A, located in intron 5, and more distant intronic substitution c.650-85A>G within intron 9 outside of the canonical splicing signals, as well as the synonymous substitutions c.372C>T (D124D) and c.1191A>C (A397A) did not produce aberrant splicing in minigene context (Fig. 1b, c). These variants are listed also in The Genome Aggregation Database (gnomAD) with a rather high allele frequency (AF): c.343-11G>A (rs143223637) AF = 0.001; c.650-85A>G (rs2075504) AF = 0.020; c.372C>T (rs140977117) AF = 0.023; and c.1191A>C (rs137923025) AF = 0.023 (http://gnomad.broadinstitute.org/). We reanalyzed DNA of the patient AKU_DB_183, who carries variant c.650-85>G, and we found already known homozygous AKU causing variants S59fs (c.175delA) in exon 3, further confirming that c.650-85A>G does not affect function.
Thus, of the eight variants with unknown significance identified in AKU-affected individuals, we demonstrated an association with exon skipping and/or cryptic/de novo splice site activation in four, c.650-13T>G, c.650-17G>A, c.650-56G>A and c.649+39T>G. This study confirms the need for combination genetic studies and in silico predictions with assays based on mRNA before concluding functional consequences of sequence alteration [40, 41].
Novel genomic deletions identified by MLPA analysis
In 10 patients, up to one HGD variant was identified by DNA sequencing. MLPA analyses of the HGD gene revealed a novel deletion of exons 5 and 6 in a patient from the Netherlands (P48), a larger deletion of exons 1, 2, 3, and 4 in a case from Germany/Peru (P25), and a deletion of exon 13 in three patients from Italy (P44, P04, P09) (S1 Table). All deletions were heterozygous and hence not seen by DNA sequencing. Deletion breakpoints will be defined. Using MLPA analysis, we were able to identify a previously reported deletion of exon 2 [42] in four patients from our cohort (P08, P16, P129, P130), as well as a novel deletion of almost the entire intron 2, originally identified by sequencing in the SONIA2 patient from Jordan (P121) (S1 Table). We plan to prepare a new MLPA probe for the in13ex14 deletion found in the patient from Iran (P15).
No copy number variation was observed in one remaining case, who showed only a single-copy HGD variant (P35, c.550-2A>C, S1 Table). This patient carried also an intronic variant ivs4+31A>G (c.282+31A>G), previously described as a benign polymorphism (HGD gene mutation database; gnomAD rs1800722 with AF 0.020) and by HSF predicted to have probably no impact on splicing (data not shown).
Predicted effect of missense variants on protein function
Localization of the 15 novel missense variants within the HGD hexamer can be seen in Figure S1. In accordance with our previous study [13], mCSM and DUET provided a structural understanding behind the inactivation of HGD activity; variants were classified based on the predicted effects into three classes: (i) those that may alter the active site, reducing activity (Active site disruption); (ii) those that destabilize the protomer, reducing activity (Protomer destabilization); and (iii) those that prevent formation of the homohexamer, disrupting activity (Hexamer disruption) (Table 1, S2 Table).
The first class of variants are predicted to affect HGD activity through direct alteration of the active site. The novel variants R330S, P332R, R347P, and Y350C were assigned to this variant effect class, based on their proximity to active site residues, their effect on binding of the substrate as determined by mCSM-Lig and upon visual inspection of the residue environment on the structure. Figure 2a, b depict the noncovalent interactions on the wild-type HGD structure established by the residues Y350 and P332, respectively, calculated using Arpeggio [43]. Y350 forms a π–π interaction with H349 as well as a main-chain hydrogen bond. Change to cysteine results in the loss of this interaction, affecting the nearby catalytic region (as shown by the catalytic iron ion and coordinating residues in Fig. 2a). Change from P332 to arginine would also affect the neighboring catalytic site (interaction network of wild-type residue depicted in Fig. 2b) given its proximity and residue environment composition (a proline-rich region). Substitution to arginine would also potentially create steric clashes, inducing conformational changes for its accommodation.
Noncovalent interaction network of residues involved in variants that disrupt HGD function by different mechanisms: (a, b) directly affecting active site (Y350 and P332); (c, d) affecting protomer stability (W97 and G205); and (e, f) affecting protein–protein affinity and hemaxer formation (R336 and G185). Hydrogen bonds are depicted as red dashes, ionic interaction as yellow dashes, π interactions as gray disks, and other polar interactions as gray dashes. Interface residues (from a different chain) are colored in dark gray
The second group of variants affects the production of active HGD enzyme through destabilization of the protomer structure. These variants typically perturb the local structure by the introduction of energetically unfavorable changes, disrupting the interactions made by the wild-type residues. Novel variants on this variant effect class, as predicted by mCSM-Stability and DUET and validated via visual inspection, include W97C, G205V, A267V, and I346T. The noncovalent interaction network on the wild-type structure for residues W97 and G205 are depicted in Fig. 2c and d [43]. W97 is inserted in a predominantly hydrophobic environment and performs a series of carbon–π and amide–π interactions with neighboring residues (Fig. 2c), which would be disrupted by change to cysteine, as predicted by DUET (ΔΔG = −1.22 kcal/mol), destabilizing the protomer. G205 is surrounded by polar/charged residues and forms a main-chain to main-chain hydrogen bond. Substitution to valine with its hydrophobic side chain would be unfavorable. Additionally, G205 is adopting a positive phi (φ) conformation, which would also be inaccessible for larger residues.
The variants in the third group are located at the interfaces between protomers and are likely to affect the enzyme activity by lowering stability or preventing formation of the symmetrical homohexameric structure. Novel variants that fall into this class include S150L, D18Y, G152R, G170A/S, G185R, and R336T. Substitution of R336 to threonine was predicted by mCSM-PPI to highly destabilize protein–protein affinity (ΔΔG = −1.59 kcal/mol). As depicted in Fig. 2e, R336 established several interchain interactions, including a hydrogen bond to G185 of a different chain, and a series of ionic interactions with two aspartic acids (E168 and E42). Change to threonine would result in the loss of these interactions, greatly impacting protein–protein affinity and, therefore, hexamer formation. G185 is also sitting on the protein–protein interface. Change to arginine is predicted to be highly detrimental to hexamer formation by mCSM-PPI (ΔΔG = −2.20 kcal/mol), as the residue is on a loop and adopting a positive phi (φ) conformation, most likely inaccessible to an arginine on that environment. Substitution to a larger residue would lead to steric clashes.
In addition, we analyzed the effect of a novel insertion A218_N219insKI on the HGD structure. This insertion is located on a loop at the interface between the two stacked trimers, interacting with two protomers from the other trimeric unit. The insertion would disrupt these interactions and prevent formation of the hexameric complex.
A similar analysis was performed for all 111 variants described in the HGD gene so far (found at 93 distinct amino acid residue positions within the structure, including the known polymorphism H80Q), and their localization was analyzed within a general structure of the HGD hexamer, employing static and dynamic methods (S2 Table).
Conservation of 93 mutated residues currently described in HGD
The Shannon entropy value (H) reflects the variability of every residue between compared sequences, and consequently, its conservation. Using Bio3D [24] analysis of 1000 aligned HGDHs homologous proteins, we obtained evaluation of the evolutionary conservation for all 445 HGDHs amino acid residues. Complementary Shannon entropy H.10 values for the 93 residue positions affected by the 111 known missense changes are shown in S2 Table, and for new variants also in Table 1. As shown in Fig. 3a, the 93 mutated residues in AKU tend to be the most highly conserved, missense, mutation-intolerant residues. In addition, as Fig. 3b illustrates, missense variants that affect function were generally associated with highly conserved, low entropy values (207 amino acid positions within HGD, which have H.10 of 0.0–0.2).
Position of missense variants within HGD monomer and their conservation. (a) Position of 111 missense variants within HGD protomer shows that the 93 residues which are mutated in AKU (shown as spheres) tend to be the most highly conserved, missense variant intolerant residues (blue conserved/intolerant—red variable/tolerant). (b) The incidence of missense variants (y-axis) at the positions with different Shannon entropy rates (x-axis) divided into intervals of 0.1. Solid line (“All”) represents values calculated on the total number of positions; dotted line (“mut”) are values calculated on the 93 positions that carry one or more variants. The graph can be divided into three regions: (i) H.10 between 0 and 0.2: positions with high probability of pathogenic variants affecting function; it includes highly conserved residues, which are hit by the large part of missense variants; (ii) H.10 between 0.2 and 0.8: positions with high probability to be affected by variants compromising function and need to be analyzed in each individual case; (iii) H.10 between 0.8 and 1: predicted variants-free positions. (c) MTR (missense tolerance ratio) plot for HGD. MTR is a new scoring system describing population-based observations as opposed to homolog-based alignments. Regions in red achieved a study-wide FDR < 0.05. MTR = 1, depicted by the blue dashed line. Multiple gene-specific estimates are also depicted, including a gene’s median MTR (black dashed line), 25th percentile MTR (dark green dashed line), and 5th percentile lowest MTR estimates (orange dashed line). The majority of pathogenic genetic disease variants normally fall below the 25th percentile
Higher Shannon entropy values, coinciding with highly variable residues, were less likely to be associated with variants affecting function. An increase in the missense variant incidence (dotted line) for values of H.10 between 0.8 and 1.0 (region of high variability, thus predictive of mutation-free positions) is caused by missense variants R347P (one of the novel variants) and E143D. After analyzing position 143 by Bio3D [24], we could see a similar incidence of glutamic acid and aspartic acid residues at this position in the course of evolution (S3 Table). This led us to hypothesize that E143D is likely to be a polymorphism rather than a pathologic variant. R347P, however, affects a position near the active site, and proline occurred very infrequently at this position across the 1000 identified homologs (S3 Table), which suggests that the novel R347P variant affects HGD function.
Bio3D analysis was also extended to the variants with H.10 entropy values between 0.2 and 0.8 (positions with high probability to be affected by variants with effect on function, but requiring individual analysis) (S4 Table). The only discordance was noticed for F169L and H80Q, which is a known HGD polymorphism in exon 4. Leucine at position 169 has a substitution frequency of 0.35. In order to explain the effect of this change on the protein function, a combined analysis of the folding landscape and sequence coevolution has been performed (data not shown). This confirms the F169L variant as deleterious in spite of conflicting results from prediction tools [38]. Moreover, it suggests its destabilizing role to be exerted through hexamer assembly disruption rather than protomer folding destabilization.
In addition to assessing conservation across evolution, we also considered population-based variability using the MTR scoring system [25]. The majority of HGD variants affecting function had an MTR score in the bottom 25th percentile, reflecting their intolerance to incorporating variation at that site (Fig. 3c).
HGD variants within the general HGD structure
The HGD protein can be divided into four general structural sections: core, active site, surface, and homohexamer interfaces. Further examination of the hydrophobic interactions revealed that the core could be divided into two regions (Figure S2A), showing that each protomer is formed by two structural domains, one of which fully contains the active site. In the HGD hexameric structure, the surface residues were mainly hydrophilic, as expected of globular proteins (Figure S2C). In addition, we confirmed the presence of a pore within the protomer structure [44], with a channel, in which the side chains of a large number of residues are exposed (Figure S2B). Most likely, these residues could be directly or indirectly involved in the complex HGD catalytic function and could represent critical points in which a missense variant would lead to an alteration of the active site functionality. Therefore, we refer to a common category Active site/Pore in the S2 Table, which indicates for each known HGD missense variant the involved structural region, as well. However, as can be seen in Fig. 4, only the surface category shows a variant incidence lower than the overall, which might indicate that within a complex structure of the HGD hexamer the missense variants are better tolerated if surface residues are affected.
Frequency of the variants affecting different structural regions within HGD protein: the number of the mutated positions within the specific region was divided by the number of all residues that represent given region. The overall variant incidence was calculated by dividing the total number of positions mutated (93) by total number of HGD residues (445 AA). Each structural regions is characterized by aminoacidic properties
The amino acid composition analysis of each group (Fig. 4) shows how Interface and Surface groups have a similar trend as overall, while core and active site/pore display some specific features: the core structural group has only two amino acid categories, hydrophobic (typical amino acid residues found in the core of protein structures), and Proline; while in the active site/pore we can observe the absence of polar amino acids and glycines and a high percentage of charged positive amino acid residues.
S5 Table shows the AKU variants propensity of different categories of each region to change into others. For the interface region, negatively charged residues showed a 60% propensity to mutate to positively charged residues; while within the surface region, 75% of polar and 60% of glycine residues mutate to positively charged and hydrophobic residues, respectively. Interestingly, in the core region, hydrophobic residues mutate to another hydrophobic ones in most cases (75%). At the active site/pore, the most prevalent, charged positive residues mutate to some other charged positive residue or into polar ones (40% each). However, the number of residues involved in most of categories is too small to allow making general conclusions.
Molecular dynamics analysis of inter- trimeric and intra-trimeric interactions
Protomer–protomer interactions in the hexameric structure are strictly noncovalent. The averaged contact map (see Figure S3) computed over the MD simulation trajectory with suitable cut-off accounts for all types of interaction from van der Waals up to long-range electrostatics and gives both a detailed list of residue pairing, as well as an overview on subunit interaction. Specifically, taking subunit A as a reference, interactions were found with: (a) residues within the trimer (intra-trimer), the N-terminus domain of the chain A binds the C-terminus domain of chain B, and the C-terminus domain of chain A binds N-terminus domain of the subunit C; (b) residues of the other trimers (inter-trimer): the N-terminus of the subunit A interacts with the N-terminus of chain D, and the C-terminus domain with the C-terminus of subunit F. The only noninteracting pair is chain A/chain E. Hydrogen-bond stability was also evaluated by computing their occupancy along the MD simulation. The existence of a stable H-bond and/or other noncovalent bonding between subunits was taken as a criterion to assign the involved amino acid residues to the “interface residues” group; see Figure S2D for a graphical overview and the corresponding column of S2 Table for correlation with missense variants. The same table shows good agreement between interface identity and hexamer disruption predicted by mCSM.
Molecular dynamics simulation of recurrent variants M368V, G161R, and A122V
An attempt to evaluate the effects of three common variants, M368V, G161R, and A122V, on structural stability by MD simulation was also carried out. Analyses of the MD trajectories of the mutated HGDHs structures by rmsd, rmsf, radius of gyration, h-bonds, contact map, and total surface access area were performed, and results compared to those of the wild type. Little difference was observed between the wild-type and the mutants. The possibility that the variants exert a destabilizing effect on an earlier structural intermediate of the protomer or hexamer cannot be excluded. However, mCSM predicted that the variants A122V and M368V would lead to disruption of the hexamer, while G161R would destabilize the protomer (S2 Table). The large and tight hexameric structure of HGD possibly makes MD simulation unsuitable for a fast and extended evaluation of the many variants affecting the enzyme, in favor of the much more high-throughput method mCSM.
Functional analysis of the variants G161R, M368V, and A122V
To experimentally assess the effects of the three most frequent variants, we performed a detailed functional analysis. All three mutants had significantly reduced activity compared to the wild-type enzyme, ranging from <1% of wild-type activity for G161R, to 31% and 34% for A122V and M368V, respectively. This is consistent with previous in vitro studies on mutants, which showed that some single residue substitutions retained substantial activity, with catalytic efficiencies (estimated as Vmax/Km) in the range of 7–25% of the wild-type [30].
We then looked at the molecular weight of the complexes using SEC-MALS, which showed that the A122V and M368V variants had <10% of the protein at a molecular weight consistent with the hexameric form. By comparison, over 90% of both the wild type and G161R variants had a mass that corresponded to the hexameric form. Despite this, the G161R variant had a thermal melting point 9 °C lower than the wild type. This is consistent with the mCSM predictions that A122V and M368V disrupt the activity of HGD through destabilization of the oligomeric complex, whilst G161R disrupts activity through destabilization of the protomer.
Many AKU patients are compound heterozygotes for two different missense variants. In such cases, the role of each missense variant is not clear, since the hexamer could be assembled with monomers all affected by the same variant (homo-oligomer) or by two different ones (hetero-oligomer) [45]. The destructive effects of variants affecting two different regions could be additive, whereas for the ones belonging to the same region, the effects could potentially compensate for each other. So far, we do not have tools to evaluate such events.
Correlations of the genotype data with serum and urine HGA, and with clinical symptoms, in patients in the SONIA2 study
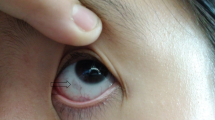
Theoretically, the differences in residual catalytic activity of the HGD proteins carrying different variants can lead to different amounts of nonmetabolized HGA. This in turn could result in differences in serum levels of HGA, as well as differences in amounts excreted in the urine, and consequently differences in disease severity. We compared both serum levels and urinary excretion of HGA in patients who are homozygous for the G161R variant (1% residual HGD activity, Group A) to those who are homozygous for the M368V or A122V variants (>30% residual HGD activity, Group B). The u-HGA/u-urea ratio was statistically significantly (p = 0.037) higher in Group A compared to Group B, with mean (SD) values of 124 (22.7) and 107 (21.6), respectively, indicating that Group B has retained some ability to metabolize HGA also in vivo. There was, however, no difference in s-HGA or u-HGA24 between the two groups (Table 2). This indicates that other factors, mainly the amount of dietary intake of protein (i.e., of tyrosine) but also the patient’s renal function [46], is more important for circulating concentrations of HGA, and amounts excreted in urine, than differences in residual HGD activity. There was no difference in eye pigmentation, bone density, or degree of scoliosis between the two groups, while peak aortic velocity was significantly higher (p = 0.024) in Group A (1.95 [0.787] m/s) than in Group B (1.45 [0.358]). This difference is most likely explained by the difference in age in the two groups, rather than by differences in HGA levels. Group A was 52.8 (8.74) years old, and Group B 41.5 (7.75) years old (p < 0.001, Table 2).
When looking at data for all SONIA2 patients, there is a marked increase in peak aortic velocity, a measure of possible aortic sclerosis or stenosis, from about age 45 to 50 years (data not shown). It should also be kept in mind that in both Groups A and B, u-HGA24 is on average more than 10,000 times higher than normal, i.e., >30 mmol in the patients compared to <3 µmol in non-AKU subjects [47], while the difference in u-HGA/u-urea between Groups A and B is only about 15%. Thus, even in Group B, HGA levels are very high and other factors than genotype will affect the many clinical manifestations of AKU.
However, understanding the effect of each variant on the HGD structure and function and its possible residual activity can have implications for the development of possible novel treatment strategies for selected variants in AKU. Missense variants in particular seem to be a good target for approaches aiming at a total or partial rescue of enzyme activity by targeting the HGD with pharmacological chaperones, i.e., small molecules helping structural stability [48].
Conclusions
We present the systematic analysis of the largest cohort of patients from 39 countries with the iconic rare metabolic disorder AKU. We identified 28 novel HGD gene variants, including three novel larger genomic deletions identified by MLPA, which was performed for the first time in AKU. Four of eight variants predicted to affect splicing were shown to cause exon skipping and/or cryptic/de novo splice site activation. In summary, using DNA sequencing, MLPA, and splicing minigene reporter assay, we were able to identify AKU-causing variant in 343 of 344 AKU chromosomes (99.7%).
In one patient, only one HGD variant was found by sequencing and MLPA analysis. Deep intronic HGD variants affecting correct exon splicing might represent an alternative mutation mechanism in these cases. However, such variants can be identified by the analysis of the patient’s cDNA extracted from the tissues expressing HGD, such as liver, kidney, or cartilage, which was not available.
AKU is normally characterized through genetic changes in the HGD gene but the identification of variants likely affecting structure is not always straightforward. Evolutionary conservation (Shannon entropy) and population conservation (MTR) scores indicated that AKU variants were located at more conserved residue positions. This could provide insight into novel missense variants that have a high probability of being deleterious.
Structural analysis of the AKU causal variants revealed that AKU causal missense variants were likely to lead to changes in protein folding and stability, or interactions with other protomers or substrate. mCSM-Stability, mCSM-PPI, and mCSM-Lig predictions were able to effectively differentiate AKU causal missense variants from nonpathogenic, and could prove to be a quick, direct, and effective tool to identify missense variants that could compromise enzyme activity [49, 50].
For the three most frequent missense variants, the mCSM predictions correlated well with the experimental data, obtained by studying expressed and purified AKU mutants. By contrast, our molecular dynamics analysis of these variants was not sensitive enough to uncover their effect in the context of the complex structure of the HGD hexamer.
We observed a small, but not clinically relevant, difference in the u-HGA/u-urea ratio between patients who are homozygous for the variant with almost no residual HGD activity (G161R) and patients with a >30% residual activity (M368V or A122V). Absolute urinary excretion (u-HGA24) and serum concentrations of HGA were, however, not different in the two groups, indicating that dietary protein intake is more relevant for how much nonmetabolized HGA accumulates in the body than is the genotype. There was also no difference in severity in the tested AKU symptoms, except for peak aortic velocity, which was higher in the group with low HGD activity. This difference is most likely explained by the fact that this group also consisted of older patients than the group with higher enzyme activity, not by the relatively small differences in HGA levels in the two groups.
Understanding the effect of each variant on the HGD structure and function and its possible residual activity can have implications, in case of suitable variants, for a development of new possible treatment strategies, i.e., use of small molecules to help structural stability in order to totally or partially rescue enzyme activity.
References
Garrod AE. Croonian lectures on inborn errors of metabolism, lecture II: alkaptonuria. Lancet. 1908;2:73–79.
La DuBN, Zannoni VG, Laster L, Seegmiller JE. The nature of the defect in tyrosine metabolism in alcaptonuria. J Biol Chem. 1958;230:251–60.
Al-Sbou M, Mwafi N. Nine cases of Alkaptonuria in one family in southern Jordan. Rheumatol Int. 2012;32:621–5.
Janocha S, Wolz W, Srsen S, et al. The human gene for alkaptonuria (AKU) maps to chromosome 3q. Genomics. 1994;19:5–8.
Sakthivel S, Zatkova A, Nemethova M, Surovy M, Kadasi L, Saravanan MP. Mutation screening of the HGD gene identifies a novel alkaptonuria mutation with significant founder effect and high prevalence. Ann Hum Genet. 2014;78:155–64.
Srsen S, Varga F. Screening for alkaptonuria in the newborn in Slovakia. Lancet. 1978;2:576.
Zannoni VG, Lomtevas N, Goldfinger S. Oxidation of homogentisic acid to ochronotic pigment in connective tissue. Biochim Biophys Acta. 1969;177:94–105.
Ranganath LR, Jarvis JC, Gallagher JA. Recent advances in management of alkaptonuria (invited review; best practice article). J Clin Pathol. 2013;66:367–73.
Pollak MR, Chou YH, Cerda JJ, et al. Homozygosity mapping of the gene for alkaptonuria to chromosome 3q2. Nat Genet. 1993;5:201–4.
Fernández-Cañón JM, Granadino B, Beltrán-Valero de Bernabé D, et al. The molecular basis of alkaptonuria. Nat Genet. 1996;14:19–24.
Fernández-Cañón JM, Peñalva MA. Molecular characterization of a gene encoding a homogentisate dioxygenase from Aspergillus nidulans and identification of its human and plant homologues. J Biol Chem. 1995;270:21199–205.
Granadino B, Beltrán-Valero de Bernabé D, Fernández-Cañón JM, Peñalva MA, Rodríguez, de Córdoba S. The human homogentisate 1,2-dioxygenase (HGO) gene. Genomics. 1997;43:115–22.
Nemethova M, Radvanszky J, Kadasi L, et al. Twelve novel HGD gene variants identified in 99 alkaptonuria patients: focus on ‘black bone disease’ in Italy. Eur J Hum Genet. 2016;24:66–72.
Zatkova A, Sedlackova T, Radvansky J, et al. Identification of 11 novel homogentisate 1,2 dioxygenase variants in Alkaptonuria patients and establishment of a novel LOVD-based HGD mutation database. JIMD Rep. 2012;4:55–65.
Laschi M, Tinti L, Braconi D, et al. Homogentisate 1,2 dioxygenase is expressed in human osteoarticular cells: implications in alkaptonuria. J Cell Physiol. 2012;227:3254–7.
Titus GP, Mueller HA, Burgner J, Rodríguez de Córdoba S, Peñalva MA, Timm DE. Crystal structure of human homogentisate dioxygenase. Nat Struct Biol. 2000;7:542–6.
Zatkova A. An update on molecular genetics of Alkaptonuria (AKU). J Inherit Metab Dis. 2011;34:1127–36.
Ranganath LR, Milan AM, Hughes AT, et al. Suitability Of Nitisinone In Alkaptonuria 1 (SONIA 1): an international, multicentre, randomised, open-label, no-treatment controlled, parallel-group, dose-response study to investigate the effect of once daily nitisinone on 24-h urinary homogentisic acid excretion in patients with alkaptonuria after 4 weeks of treatment. Ann Rheum Dis. 2016;75:362–7.
Olsson B, Cox TF, Psarelli EE, et al. Relationship between serum concentrations of nitisinone and its effect on homogentisic acid and tyrosine in patients with Alkaptonuria. JIMD Rep. 2015;24:21–27.
Spiga O, Cicaloni V, Bernini A, Zatkova A, Santucci A. ApreciseKUre: an approach of precision medicine in a rare disease. BMC Med Inform Decis Mak. 2017;17:42.
den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12.
Kralovicova J, Knut M, Cross NC, Vorechovsky I. Identification of U2AF(35)-dependent exons by RNA-Seq reveals a link between 3’ splice-site organization and activity of U2AF-related proteins. Nucleic Acids Res. 2015;43:3747–63.
Kralovicova J, Gaunt TR, Rodriguez S, Wood PJ, Day IN, Vorechovsky I. Variants in the human insulin gene that affect pre-mRNA splicing: is -23HphI a functional single nucleotide polymorphism at IDDM2? Diabetes. 2006;55:260–4.
Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–6.
Traynelis J, Silk M, Wang Q, et al. Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res. 2017;27:1715–29.
Joosten RP, te Beek TA, Krieger E, et al. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011;39:D411–419.
Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42.
Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comp Phys Comm. 1995;91:43–56.
DeLano WL. PyMOL(TM) molecular graphics system, version 1.7.2.1. 2014.
Rodriguez JM, Timm DE, Titus GP, et al. Structural and functional analysis of mutations in alkaptonuria. Hum Mol Genet. 2000;9:2341–50.
Chan LJ, Ascher DB, Yadav R, et al. Conjugation of 10 kDa linear PEG onto Trastuzumab Fab’ is sufficient to significantly enhance lymphatic exposure while preserving in vitro biological activity. Mol Pharm. 2016;13:1229–41.
Chan LJ, Bulitta JB, Ascher DB, et al. PEGylation does not significantly change the initial intravenous or subcutaneous pharmacokinetics or lymphatic exposure of trastuzumab in rats but increases plasma clearance after subcutaneous administration. Mol Pharm. 2015;12:794–809.
Hermans SJ, Ascher DB, Hancock NC, et al. Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides. Protein Sci. 2015;24:190–9.
Pacitto A, Ascher DB, Wong LH, et al. Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open Biol. 2015;5:150174.
Hughes AT, Milan AM, Christensen P, et al. Urine homogentisic acid and tyrosine: simultaneous analysis by liquid chromatography tandem mass spectrometry. J Chromatogr B. 2014;963:106–12.
Hughes AT, Milan AM, Davison AS, et al. Serum markers in alkaptonuria: simultaneous analysis of homogentisic acid, tyrosine and nitisinone by liquid chromatography tandem mass spectrometry. Ann Clin Biochem. 2015;52:597–605.
Beltrán-Valero de Bernabé D, Granadino B, Chiarelli I, et al. Mutation and polymorphism analysis of the human homogentisate 1, 2-dioxygenase gene in alkaptonuria patients. Am J Hum Genet. 1998;62:776–84.
Usher JL, Ascher DB, Pires DE, Milan AM, Blundell TL, Ranganath LR. Analysis of HGD gene mutations in patients with Alkaptonuria from the United Kingdom: identification of novel mutations. JIMD Rep. 2015;24:3–11.
Kralovicova J, Christensen MB, Vorechovsky I. Biased exon/intron distribution of cryptic and de novo 3’ splice sites. Nucleic Acids Res. 2005;33:4882–98.
Grodecka L, Buratti E, Freiberger T. Mutations of pre-mRNA splicing regulatory elements: are predictions moving forward to clinical diagnostics? Int J Mol Sci. 2017;18:1668.
Raponi M, Kralovicova J, Copson E, et al. Prediction of single-nucleotide substitutions that result in exon skipping: identification of a splicing silencer in BRCA1 exon 6. Hum Mutat. 2011;32:436–44.
Zouheir Habbal M, Bou-Assi T, Zhu J, Owen R, Chehab FF. First report of a deletion encompassing an entire exon in the homogentisate 1,2-dioxygenase gene causing alkaptonuria. PLoS One. 2014;9:e106948.
Jubb HC, Higueruelo AP, Ochoa-Montano B, Pitt WR, Ascher DB, Blundell TL. Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J Mol Biol. 2017;429:365–71.
Jeoung J-HBM, Lin T-Y, Dobbek H:. Visualizing the substrate-, superoxo-, alkylperoxo-, and product-bound states at the nonheme Fe(II) site of homogentisate dioxygenase. Proc Natl Acad Sci USA. 2013;110:12625–30.
Gupta V, Kalaiarasan P, Faheem M, Singh N, Iqbal MA, Bamezai RN. Dominant negative mutations affect oligomerization of human pyruvate kinase M2 isozyme and promote cellular growth and polyploidy. J Biol Chem. 2010;285:16864–73.
Introne WJ, Phornphutkul C, Bernardini I, McLaughlin K, Fitzpatrick D, Gahl WA. Exacerbation of the ochronosis of alkaptonuria due to renal insufficiency and improvement after renal transplantation. Mol Genet Metab. 2002;77:136–42.
Davison AS, Milan AM, Hughes AT, Dutton JJ, Ranganath LR. Serum concentrations and urinary excretion of homogentisic acid and tyrosine in normal subjects. Clin Chem Lab Med. 2015;53:e81–83.
Bernini A, Galderisi S, Spiga O, et al. Toward a generalized computational workflow for exploiting transient pockets as new targets for small molecule stabilizers: application to the homogentisate 1,2-dioxygenase mutants at the base of rare disease Alkaptonuria. Comput Biol Chem. 2017;70:133–41.
Pires DE, Ascher DB, Blundell TL. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30:335–42.
Pires DE, Ascher DB, Blundell TL. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res 2014;42:W314–W319.
Acknowledgements
We would like to thank Drs. Jozef Rovensky (Piestany, Slovakia), Kim-Hanh Le Quan Sang (Paris, France), Berardino Porfirio (Florence, Italy), Caterina Aurizi (Rome, Italy), Anastasia Skouma (Athens, Greece), Pallavi Vats (New Delhi, India), and Robert Aquaron (Marseille, France) for DNA samples of AKU patients. The European Commission Seventh Framework Program funding granted in 2012 supported the SONIA1, SONIA2, and SOFIA studies and the AKU research at the Laboratory of genetics (DevelopAKUre, project number: 304985). DBA and DEVP were funded by a Newton Fund RCUK-CONFAP Grant awarded by The Medical Research Council (MRC) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) (MR/M026302/1, APQ-00828-15). DEVP received support from the Instituto René Rachou (IRR/FIOCRUZ Minas) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (409780/2016-2), Brazil. DBA was supported also by a C.J. Martin Research Fellowship from the National Health and Medical Research Council of Australia (APP1072476) and the Jack Brockhoff Foundation (JBF 4186, 2016). This work was partially supported by MIUR Progetto Dipartimenti di Eccellenza 2018–2022.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ascher, D.B., Spiga, O., Sekelska, M. et al. Homogentisate 1,2-dioxygenase (HGD) gene variants, their analysis and genotype–phenotype correlations in the largest cohort of patients with AKU. Eur J Hum Genet 27, 888–902 (2019). https://doi.org/10.1038/s41431-019-0354-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-019-0354-0
This article is cited by
-
Alkaptonuria
Nature Reviews Disease Primers (2024)
-
Homogentisate 1,2-dioxygenase (HGD) gene variants in young Egyptian patients with alkaptonuria
Scientific Reports (2023)
-
Breakpoints characterisation of the genomic deletions identified by MLPA in alkaptonuria patients
European Journal of Human Genetics (2023)
-
Bioinformatics Analysis Reveals Novel Differentially Expressed Genes Between Ectopic and Eutopic Endometrium in Women with Endometriosis
The Journal of Obstetrics and Gynecology of India (2023)
-
A robust bacterial high-throughput screening system to evaluate single nucleotide polymorphisms of human homogentisate 1,2-dioxygenase in the context of alkaptonuria
Scientific Reports (2022)