Abstract
Primary familial brain calcification (PFBC) is a rare cerebral microvascular calcifying disorder with a wide spectrum of motor, cognitive, and neuropsychiatric symptoms. It is typically inherited as an autosomal-dominant trait with four causative genes identified so far: SLC20A2, PDGFRB, PDGFB, and XPR1. Our study aimed at screening the coding regions of these genes in a series of 177 unrelated probands that fulfilled the diagnostic criteria for primary brain calcification regardless of their family history. Sequence variants were classified as pathogenic, likely pathogenic, or of uncertain significance (VUS), based on the ACMG-AMP recommendations. We identified 45 probands (25.4%) carrying either pathogenic or likely pathogenic variants (n = 34, 19.2%) or VUS (n = 11, 6.2%). SLC20A2 provided the highest contribution (16.9%), followed by XPR1 and PDGFB (3.4% each), and PDGFRB (1.7%). A total of 81.5% of carriers were symptomatic and the most recurrent symptoms were parkinsonism, cognitive impairment, and psychiatric disturbances (52.3%, 40.9%, and 38.6% of symptomatic individuals, respectively), with a wide range of age at onset (from childhood to 81 years). While the pathogenic and likely pathogenic variants identified in this study can be used for genetic counseling, the VUS will require additional evidence, such as recurrence in unrelated patients, in order to be classified as pathogenic.
Similar content being viewed by others
Introduction
Primary familial brain calcification (PFBC) is a rare neuropsychiatric disorder characterized by abnormal calcium–phosphate deposits in the microvessels of the basal ganglia and other brain regions. Clinical manifestations can start at any age (median 31 years, range 6–77 years) [1], and include a wide spectrum of movement disorders (dystonia, parkinsonism, tremor, and chorea), neuropsychiatric symptoms (behavioral disturbances, psychosis, mood disorder, and cognitive impairment), cerebellar signs, and other symptoms [2], while up to 42% of the patients remain asymptomatic [1]. Even though the clinical presentation is variable, the neuroradiological picture (evidence of bilateral calcification affecting at least the basal ganglia) is thought to be invariably present by the age of 50. Hence, the diagnosis relies on a computerized tomography (CT) scan, in the absence of other known causes of brain calcification [2]. PFBC is typically inherited as an autosomal-dominant trait, and to date four causative genes have been identified.
SLC20A2 (solute carrier family 20, member 2) was the first gene to be linked to PFBC [3]. Since its discovery, many protein-truncating and deleterious missense variants have been identified, accounting for up to 40% of the familial cases [4]. SLC20A2 encodes the transmembrane sodium-inorganic phosphate cotransporter PiT2, suggested to have a role in phosphate clearance from the cerebrospinal fluid by recent in vitro and knockout mice studies [5].
Variants in the PDGFRB gene [6,7,8], encoding the platelet-derived growth factor receptor β (PDGF-Rβ), and in the PDGFB gene (PDGF-Rβ’s main ligand) [9,10,11,12], have been reported in more than 20 unrelated probands so far. PDGFB–PDGF-Rβ signaling mediates survival, differentiation, and migration of mesenchymal cells, including the vascular smooth muscle cells affected by calcifications in PFBC [13]. While increased signaling is associated with cancers, overgrowth, and progeria syndromes [14,15,16,17,18], in PFBC patients, protein-truncating PDGFB and missense PDGFB and PDGFRB variants lead to decreased PDGFB–PDGF-Rβ signaling [8, 19, 20]. Although PDGFB–PDGF-Rβ signaling is implicated in the regulation of inorganic phosphate transport [21], the mechanisms leading to microvascular calcification remain unknown [19].
More recently, missense variants in another phosphate transporter, encoded by the XPR1 gene, were identified in several PFBC families [22]. Subsequent functional studies showed that XPR1 mutant proteins had severely reduced membrane localization and/or impaired phosphate efflux activity [22, 23].
The interpretation of sequence variants identified in genetic screens for rare diseases remains challenging. The American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG-AMP) recently established a set of guidelines to classify genetic variants into five categories from benign (1) to pathogenic (5) [24]. While large sequence variant databases, such as gnomAD [25], are helpful in estimating allele frequencies in control populations, for rare diseases with incomplete penetrance (such as PFBC), variant recurrence in unrelated patients and family segregation data remain critical for interpretation.
In an international effort, four centers from France, USA, Italy, and Brazil gathered and analyzed sequence data from the four genes known to cause autosomal-dominant PFBC.
Materials and methods
Patients
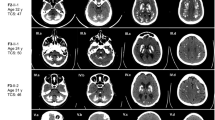
We included patients with brain calcification who were referred to four centers of expertise: University of California, Los Angeles, USA; IRCCS Neurological Institute C. Besta, Milan, Italy; Inserm U1245, Rouen, France; and Universidade Federal de Pernambuco, Recife, Brazil. All patients presented calcifications affecting at least both lenticular nuclei, beyond the age-specific severity threshold [7], a normal phospho-calcic assessment (including at least calcium, phosphate, and PTH) in blood, and no other known etiology. Probands and, if available, family members underwent clinical examination and blood sampling. Details on clinical and family history were obtained by direct interview and/or by reviewing medical records. All individuals included in this study had a brain CT scan; for some, however, details about the extent and localization of brain calcifications were not available. Detailed inclusion criteria are reported in Supplementary Methods. All participants signed written informed consent for genetic analyses.
Genetic screening
Genomic DNA was extracted from peripheral blood by standard methods. For samples from the French, US, and Brazilian series, PCR amplification and subsequent Sanger sequencing of all protein-coding exons and exon–intron boundaries of SLC20A2, PDGFB, PDGFRB, and XPR1 genes was performed as previously described [3, 6, 9, 22]. All 49 patients from the Italian series were screened with a customized gene panel (Nextera Rapid Capture Custom Enrichment), which included the PFBC genes and 55 additional genes responsible for diseases characterized by cerebral calcification (Supplementary Methods). The following genomic and transcript references were used for variant nomenclature and exon numbering: NG_032161.1 and NM_006749.4 for SLC20A2, NG_012111.1 and NM_002608.2 for PDGFB, NG_023367.1 and NM_002609.3 for PDGFRB, and NG_050964.1 and NM_004736.3 for XPR1.
Copy-number variation
Quantitative multiplex PCR of short fluorescent fragments (QMPSF) was used to assess the presence of copy-number variations (CNVs) encompassing SLC20A2 and PDGFB, in the French and Brazilian series, as previously described [12, 26]. For the US series, CNVs were genotyped using TaqMan copy-number assays, following the manufacturer’s instructions. Commercially available assays for the SLC20A2 (Hs00279506_cn, Hs00383415_cn), PDGFB (Hs00902096_cn and Hs01735391_cn), and PDGFRB (Hs01615581_cn, Hs02279533_cn, and Hs02258542_cn) genes were used. For the Italian series, the cn.MOPS tool was applied to next-generation sequencing data for CNV detection [27].
Variant assessment
Variant classification was conducted following ACMG-AMP recommendations [24]. Briefly, these criteria included prior identification as a PFBC-causing variant (reported in the literature, HGMD, Clinvar, and/or the PFBC variant database https://coppolalab.ucla.edu/lovd/genes), allele frequency in population databases (gnomAD [25], http://gnomad.broadinstitute.org/), computational and predictive data (Polyphen2, SIFT, MutationTaster, and splicing predictions provided by the Alamut visual software (Interactive Biosoftware, Rouen, France)), functional studies (reported in the literature), and segregation data. Each variant was first classified into one of the five ACMG-AMP classes by an investigator from the group where it was identified and then reviewed by the entire study group. All variants reported in this study were added to the PFBC database at https://coppolalab.ucla.edu/lovd/genes.
Affected relatives
Clinical and imaging data from affected relatives were collected, and genetic testing was performed on available DNA samples to ascertain variant cosegregation.
Results
Genetic screening in four series
By screening the four known PFBC-causative genes in 177 unrelated probands from four independent international series, we identified 34 probands (19.2%) carrying a variant classified as pathogenic (class 5) or likely pathogenic (class 4), while 11 carried a variant of uncertain significance (VUS) (class 3, 6.2%). In contrast, CNV analysis did not reveal any clear large deletion or duplication in the PFBC genes screened. The overall variant detection rate was therefore 25.4% (45/177) (Supplementary Table 1). Only 2 out of the 177 unrelated probands were previously reported [23, 28]. After including 11 variant-carrying affected relative members, 56 individuals are described herein.
SLC20A2 variants
We identified 27 distinct SLC20A2 variants in 30 unrelated probands (16.9%, Table 1). Nine of these variants had previously been reported in other PFBC patients [3, 4, 6, 7, 29,30,31,32], including six missense variants for which pathogenicity was uncertain and that can now be classified as pathogenic: p.(Pro184Leu) and p.(Gly498Arg), or likely pathogenic: p.(Arg71His), p.(Asn194Ser), p.(Ser434Trp), and p.(Ala585Thr). These variants were seen in 12 of our unrelated probands, including one case already reported in the literature [28]. The remaining 18 SLC20A2 variants were novel, of which nine were protein-truncating variants (PTV) and were therefore classified as pathogenic.
Two novel likely pathogenic variants were also identified. First, an in-frame deletion of 27 nucleotides (c.1822_1848del) in exon 11 of SLC20A2 was identified in a proband and his affected father. This variant is predicted to cause a deletion of nine amino acids, p.(Ile608_Trp616del), at the C-terminal domain of Pit-2, in a transmembrane region. Second, a predicted-damaging missense variant, c.541C>T, p.(Arg181Trp) in exon 5 was identified in a patient and his affected father. This variant was found in one individual from the gnomAD database (MAF = 4.1e−06). Other missense pathogenic variants in nearby residues have been reported in PFBC patients [4], supporting evidence for pathogenicity.
Among the additional seven novel VUS identified, two were intronic (c.289+5G>A, c.290–8A>G), absent from gnomAD, and with strong in silico predictions of a splicing defect at the closest canonical site (MaxtEntScan score change of –80.7% and –54.4%, respectively, with the c.290–8A>G predicted to create a new acceptor site at position c.290–7). Two other novel missense VUS were located at exon boundaries. The c.290G>A, p.(Gly97Asp) variant, affecting the first base of exon 3, was predicted as damaging by in silico tools and to cause a slight effect in splicing (MaxEntScan score change: –7%). The c.1523G>A variant, p.(Ser508Asn), affecting the last base of exon 8, was also predicted to be damaging, in addition to a strong effect on splicing (MaxEntScan score change: –59.5%). RNA from these patients was not available to confirm the hypothesis of a protein-truncating effect through altered splicing, precluding their classification as (likely) pathogenic. The other novel VUS, p.(His488Arg), p.(Gly589Arg), and p.(Val624Glu), were not detected in gnomAD and are predicted to be damaging by in silico analysis. Even though other missense pathogenic variants in nearby residues have been reported, there was not sufficient evidence to classify these specific variants as (likely) pathogenic.
PDGFB variants
We identified six distinct PDGFB variants in 6 unrelated probands (3.4%, Table 2). Two of these variants had already been reported in other PFBC patients: nonsense p.(Arg149Ter) and, stop loss c.726G>C, p.(Ter242TyrExtTer89) that adds 89 residues to the protein [9]. We identified a novel stop loss variant, c.724T>C, p.(Ter242GlnExtTer89), which is also predicted to cause an elongation of the reading frame by 89 amino acids. Functional studies have shown that proteins with variants causing a C-terminal extension, namely p.(Ter242TyrExtTer89), failed to induce any detectable PDGF-Rβ autophosphorylation [19]. A novel canonical splice site variant, c.456+1G>A (Table 2), predicted to affect splicing of exon 4 in PDGFB, was identified in a proband and the affected mother. Both of these novel variants were absent from gnomAD. Therefore, there was enough evidence to support these variants as pathogenic for PFBC.
We also identified two novel missense variants, both absent from gnomAD and predicted damaging by in silico analysis: p.(Gly132Arg) and p.(Arg142His) (Table 2). Variant p.(Gly132Arg) was identified in an additional unrelated French patient with brain calcifications (enrolled after the data freeze, hence not included in this series) and was therefore classified as likely pathogenic.
PDGFRB variants
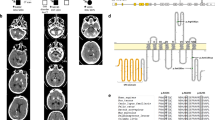
Three distinct PDGFRB variants were found in three unrelated probands (1.7%, Table 3): p.(Arg226Cys), p.(Pro596Leu), and p.(Asp844Gly), all novel missense variants, predicted damaging. Of these, only the p.(Pro596Leu) variant was present in two individuals in gnomAD (MAF = 8.1e−06). Segregation data was only available for the family carrying the p.(Asp844Gly) variant and we showed that this variant resulted in a loss of PDGFRβ autophosphorylation (Supplementary Figure 1). Based on this evidence, this variant was classified as pathogenic, while the other two were classified as VUS.
XPR1 variants
Five distinct XPR1 variants were found in six unrelated probands (3.4%, Table 4). Two of these variants had already been associated with PFBC. One of our unrelated French patients carried the same p.(Leu145Pro) variant reported in the original XPR1 paper [22]. The other variant, p.(Leu87Pro), was found in a case already reported [23]. These two variants were not found in gnomAD and can be classified as pathogenic based on published functional evidence [22, 23]. Three additional predicted-damaging missense variants were found (Table 4). While p.(Thr233Ser) was found in two unrelated PFBC individuals, it was also found in two individuals within the gnomAD database (MAF = 8.1e−06). On the other hand, both p.(Arg459Cys) and p.(Asn619Asp) were not found in gnomAD. Furthermore, for p.(Arg459Cys), the unaffected proband’s mother did not carry this variant and had a normal brain CT scan. Both p.(Thr233Ser) and p.(Arg459Cys) variants were therefore classified as likely pathogenic, while there was not sufficient evidence for p.(Asn619Asp), hence classified here as VUS.
Clinical presentation
Herein, we reported a total of 56 PFBC patients (32F; 24M), including the 45 probands that were found to carry VUS or (likely) pathogenic variants, and 11 relatives that had brain calcifications and the same variant as the proband (Fig. 1a). Detailed clinical and radiological data were available in 54/56 patients (Tables 1–4), and at the time of genetic testing, 44 (81.5%) of these were symptomatic (Fig. 1b). Mean age at clinical onset was 47.2 years (Fig. 1c) (median = 52 y, range: 3–81 y, age at onset was unknown for eight cases, including one with onset in childhood) and mean age at last examination was 57.4 years in symptomatic patients and 47.5 in asymptomatic patients. Parkinsonism (alone or combined with other clinical manifestations) was the most frequent finding, present in 23/44 (52.3%) of symptomatic patients, mostly with an akinetic–rigid presentation (Fig. 1d). Cognitive impairment was documented in 18/44 (40.9%) symptomatic cases, psychiatric disturbances (depression, psychosis, anxiety) in 17/44 (38.6%), while 11/44 (25.0%) patients had cerebellar signs. In addition, migraine was reported by 10/54 patients (18.5%); in five of these patients neurological examination was unremarkable and therefore they were considered asymptomatic.
Clinical presentation of 56 variant carriers. a Number of familial (including relatives) and sporadic cases, and b number of symptomatic and asymptomatic individuals per gene. c Distribution of age-at-onset (years) per gene (horizontal line represents the average age of onset across all 37 cases with known age-at-onset). d Frequency of main symptoms among the 44 symptomatic variant carriers.
Discussion
We screened the four known PFBC-causative genes in a series of 177 PFBC patients and identified 41 distinct variants, in a total of 45 unrelated probands. Taking into account only likely pathogenic and pathogenic variants, for which evidence is sufficient to propose genetic counseling, 34 out of the 177 (19.2%) unrelated probands carried such variants. However, the overall variant detection rate can increase up to 25.4% (45/177), if future studies provide new evidence to reclassify the VUS we found as causal. As expected, SLC20A2 showed the highest contribution with variants identified in 16.9% (30/177) of the probands, followed by XPR1 and PDGFB, each with 3.4% (6/177), and then PDGFRB with 1.7% (3/177). These rates are consistent with those reported in other French series that, similar to ours, had patients with and without known family history [33], in contrast to previous reports that showed high mutation rates in patients with a positive family history [34]. Even though we screened novel unrelated probands, we detected new but also previously reported PFBC variants, sometimes in patients originating from the same country as the original carrier. It should be noted that, based on available family information, none of the patients in our series seem to be related to any of the PFBC carriers already published in the literature.
SLC20A2 was the first PFBC-causative gene to be identified, linking cerebral inorganic phosphate metabolism to PFBC’s pathophysiology [3]. Evidence that SLC20A2 haploinsufficiency causes PFBC is strong as both PTV and total/partial deletions have been identified [3, 26, 29]. This hypothesis has been confirmed in mouse models [5, 35, 36], and by in vitro assessment of some of the missense variants [3]. In our series, including patients with positive family history and apparently sporadic cases, we confirmed SLC20A2 as the major causative gene, accounting for at least 13.0% of the cases (adding up to 16.9% when including VUS).
XPR1 was the most recent PFBC gene to be identified [22], and in our series, variants within this gene are as frequent as PDGFB variants. Pathogenicity of XPR1 variants reported to date has been ascertained based on: strong segregation [22], recurrence among unrelated patients, and/or functional data showing a defect in inorganic phosphate transport [22, 23]. Interestingly, all known pathogenic variants are located in the SPX domain of XPR1, the function of which remains uncertain. We identified three novel missense variants, all predicted damaging, but located outside the SPX domain. Functional analyses are needed to further clarify their role.
The identification of protein-truncating PDGFB variants following the identification of missense PDGFRB variants, provided the first evidence that decreased PDGFB–PDGFRβ signaling was causative of PFBC. Loss of function and missense variants, as well as a partial PDGFB deletion have been identified to date [1, 9, 12, 37], supporting haploinsufficiency as causal mechanism. Here, we report four novel variants, including one PTV, one stop loss and two missense variants, of which one could be classified as likely pathogenic.
Since the original paper identifying PDGFRB as a PFBC causal gene, only four established pathogenic PDGFRB variants have been reported in the literature. These showed strong segregation evidence [6] and/or functional evidence of a loss of protein function [8, 19, 20]. Another missense variant, p.(Glu1071Val), originally considered as VUS has since been reclassified as likely benign based on functional studies [7, 19, 20]. More recently, 2 novel variants were identified in Chinese PFBC cases: a c.3G>A variant leading to a loss of the start codon, and a missense p.(Asp737Asn) variant [38]. Although the latter variant was considered a VUS, the start loss variant could be classified as pathogenic if considered truncating, however its functional effect remains unclear as an alternative in-frame ATG codon could theoretically be used. Herein, we report 3 additional missense variants, though only one of them could be classified as pathogenic based on segregation and functional data.
The PFBC phenotypic spectrum is wide and diverse, with intra and interfamilial heterogeneity. Although some of the variants found in this study are recurrent, their low frequency precluded any genotype-phenotype correlations, and therefore we focused on all carriers. We found that 81.5% of those with clinical information available were considered symptomatic, with severity ranging from minor signs on clinical examination to severe disability. In previous reports, including a prior independant data freeze of the French PFBC series and a meta-analysis study, the proportion of symptomatic patients was indeed lower, 58 and 64%, respectively [1, 39]. Here, the relatively high proportion of symptomatic carriers is likely due to an inclusion bias, as symptomatic probands are more likely to be offered genetic screening than asymptomatic individuals and few relatives could be included in the present report (11/56 versus 35/57 in [1]). Age of onset was comparable to previous screens, with a wide range from 3 to 81 years. Consistent with previously published series, the most frequent symptoms in our series were parkinsonism (52.3% of symptomatic individuals), cognitive impairment (40.9%), and psychiatric signs (38.6%). Interestingly, 18.5% of the 54 patients with available clinical data reported migraine without atypical features, which is in the same range as the general population [40], suggesting that migraine in patients with brain calcifications may be coincidental. This ratio is consistent with those reported in an independent series and a literature review study [1, 39], while there are also reports that showed lack of segregation between brain calcification and migraine [41].
In summary, by screening the known PFBC genes in four cohorts from America and Europe, including sporadic and familial cases, we identified variants interpreted as VUS, likely pathogenic, or pathogenic in 25.4% of the 177 probands. While variants from the latter two classes can be used for genetic counseling, segregation and/or functional studies of the VUS are necessary to help clarify their role in PFBC, and therefore no presymptomatic testing can be recommended given the current level of evidence. The novel variants reported here will help with interpretation of future genetic screens of unrelated PFBC patients and provide a list of candidates for functional studies. Finally, further prospective follow-up studies in patients carrying pathogenic variants in PFBC-related genes are needed to widen our knowledge about disease course, genetic and/or environmental factors which could influence disease penetrance and progression.
References
Nicolas G,Charbonnier C,de Lemos RR, et al. Brain calcification process and phenotypes according to age and sex: lessons from SLC20A2, PDGFB, and PDGFRB mutation carriers. Am J Med Genet B Neuropsychiatr Genet.2015;168:586–94.
Ramos EM, Oliveira J, Sobrido MJ, Coppola G. Primary familial brain calcification. In: Adam MP, Ardinger HH, Pagon RA et al, editors. GeneReviews. Seattle, WA: University of Washington; 1993–2018.
Wang C, Li Y, Shi L, et al. Mutations in SLC20A2 link familial idiopathic basal ganglia calcification with phosphate homeostasis. Nat Genet. 2012;44:254–6.
Lemos RR, Ramos EM, Legati A, et al. Update and mutational analysis of SLC20A2: a major cause of primary familial brain calcification. Human Mutat. 2015;36:489–95.
Jensen N, Autzen JK, Pedersen L. Slc20a2 is critical for maintaining a physiologic inorganic phosphate level in cerebrospinal fluid. Neurogenetics. 2016;17:125–30.
Nicolas G, Pottier C, Maltete D, et al. Mutation of the PDGFRB gene as a cause of idiopathic basal ganglia calcification. Neurology. 2013;80:181–7.
Nicolas G, Pottier C, Charbonnier C, et al. Phenotypic spectrum of probable and genetically-confirmed idiopathic basal ganglia calcification. Brain. 2013;136:3395–407.
Sanchez-Contreras M, Baker MC, Finch NA, et al. Genetic screening and functional characterization of PDGFRB mutations associated with basal ganglia calcification of unknown etiology. Human Mutat. 2014;35:964–71.
Keller A, Westenberger A, Sobrido MJ, et al. Mutations in the gene encoding PDGF-B cause brain calcifications in humans and mice. Nat Genet. 2013;45:1077–82.
Keogh MJ, Pyle A, Daud D, et al. Clinical heterogeneity of primary familial brain calcification due to a novel mutation in PDGFB. Neurology. 2015;84:1818–20.
Nicolas G, Jacquin A, Thauvin-Robinet C, et al. A de novo nonsense PDGFB mutation causing idiopathic basal ganglia calcification with laryngeal dystonia. Eur J Human Genet. 2014;22:1236–8.
Nicolas G, Rovelet-Lecrux A, Pottier C, et al. PDGFB partial deletion: a new, rare mechanism causing brain calcification with leukoencephalopathy. J Mol Neurosci. 2014;53:171–5.
Fredriksson L, Li H, Eriksson U. The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004;15:197–204.
Appiah-Kubi K, Lan T, Wang Y, et al. Platelet-derived growth factor receptors (PDGFRs) fusion genes involvement in hematological malignancies. Crit Rev Oncol/Hematol. 2017;109:20–34.
Cheung YH, Gayden T, Campeau PM, et al. A recurrent PDGFRB mutation causes familial infantile myofibromatosis. Am J Hum Genet. 2013;92:996–1000.
Johnston JJ, Sanchez-Contreras MY, Keppler-Noreuil KM, et al. A point mutation in PDGFRB causes autosomal-dominant penttinen syndrome. Am J Hum Genet. 2015;97:465–74.
Martignetti JA, Tian L, Li D, et al. Mutations in PDGFRB cause autosomal-dominant infantile myofibromatosis. Am J Hum Genet. 2013;92:1001–7.
Takenouchi T, Yamaguchi Y, Tanikawa A, Kosaki R, Okano H, Kosaki K. Novel overgrowth syndrome phenotype due to recurrent de novo PDGFRB mutation. J Pediatr. 2015;166:483–6.
Vanlandewijck M, Lebouvier T, Andaloussi Mae M, et al. Functional characterization of germline mutations in PDGFB and PDGFRB in primary familial brain calcification. PLoS ONE. 2015;10:e0143407.
Arts FA, Velghe AI, Stevens M, Renauld JC, Essaghir A, Demoulin JB. Idiopathic basal ganglia calcification-associated PDGFRB mutations impair the receptor signalling. J Cell Mol Med. 2015;19:239–48.
Giachelli CM, Jono S, Shioi A, Nishizawa Y, Mori K, Morii H. Vascular calcification and inorganic phosphate. Am J Kidney Dis. 2001;38:S34–37.
Legati A, Giovannini D, Nicolas G, et al. Mutations in XPR1 cause primary familial brain calcification associated with altered phosphate export. Nat Genet. 2015;47:579–81.
Anheim M, Lopez-Sanchez U, Giovannini D, et al. XPR1 mutations are a rare cause of primary familial brain calcification. J Neurol. 2016;263:1559–64.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
David S, Ferreira J, Quenez O, et al. Identification of partial SLC20A2 deletions in primary brain calcification using whole-exome sequencing. Eur J Human Genet. 2016;24:1630–4.
Klambauer G, Schwarzbauer K, Mayr A, et al. cn.MOPS: Mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:e69.
Carecchio M, Varrasi C, Barzaghi C, et al. Phenotypic heterogeneity of movement disorders due to intracranial calcifications with or without SLC20A2 mutations. Mov Disord. 2014;29:S47.
Baker M, Strongosky AJ, Sanchez-Contreras MY, et al. SLC20A2 and THAP1 deletion in familial basal ganglia calcification with dystonia. Neurogenetics. 2014;15:23–30.
Mi TM, Mao W, Cai YN et al. Primary familial brain calcifications linked with a novel SLC20A2 gene mutation in a Chinese family. J Neurogenet. 2017;31:149–52.
Taglia I, Mignarri A, Olgiati S, et al. Primary familial brain calcification: genetic analysis and clinical spectrum. Mov Disord. 2014;29:1691–5.
Yamada M, Tanaka M, Takagi M, et al. Evaluation of SLC20A2 mutations that cause idiopathic basal ganglia calcification in Japan. Neurology. 2014;82:705–12.
Nicolas G, Richard AC, Pottier C, et al. Overall mutational spectrum of SLC20A2, PDGFB and PDGFRB in idiopathic basal ganglia calcification. Neurogenetics. 2014;15:215–6.
Hsu SC, Sears RL, Lemos RR, et al. Mutations in SLC20A2 are a major cause of familial idiopathic basal ganglia calcification. Neurogenetics. 2013;14:11–22.
Jensen N, Schroder HD, Hejbol EK, Fuchtbauer EM, de Oliveira JR, Pedersen L. Loss of function of Slc20a2 associated with familial idiopathic Basal Ganglia calcification in humans causes brain calcifications in mice. J Mol Neurosci. 2013;51:994–9.
Wallingford MC, Chia JJ, Leaf EM, et al. SLC20A2 deficiency in mice leads to elevated phosphate levels in cerbrospinal fluid and glymphatic pathway-associated arteriolar calcification, and recapitulates human idiopathic basal ganglia calcification. Brain Pathol. 2017;27:64–76.
Yao XP, Wang C, Su HZ et al. Mutation screening of PDGFB gene in Chinese population with primary familial brain calcification. Gene. 2016 pii: S0378-1119(16)30826-5. https://doi.org/10.1002/humu.23118.
Wang C, Yao XP, Chen HT, et al. Novel mutations of PDGFRB cause primary familial brain calcification in Chinese families. J Human Genet. 2017;62:697–701.
Tadic V, Westenberger A, Domingo A, Alvarez-Fischer D, Klein C, Kasten M. Primary familial brain calcification with known gene mutations: a systematic review and challenges of phenotypic characterization. JAMA Neurol. 2015;72:460–7.
Lipton RB, Bigal ME. Migraine: Epidemiology, impact, and risk factors for progression. Headache. 2005;45(Suppl 1):S3–S13.
Ferreira JB, Pimentel L, Keasey MP, et al. First report of a de novo mutation at SLC20A2 in a patient with brain calcification. J Mol Neurosci. 2014;54:748–51.
Acknowledgements
We thank all the patients for participating in this study and the physicians who referred patients for the study. The coinvestigators of the French PFBC study group are listed in supplementary information.
Funding
This study was cosupported by CNR-MAJ, Inserm, European Union and Région Normandie. Europe gets involved in Normandie with European Regional Development Fund (ERDF) (French group). This study received support from FACEPE, CAPES, and CNPq (310150/2016-7,480255/2013-0, 440770/2016-5; IBPG-0627-2.02/11; IBPG-0162-2.02/14; BFP-0123-2.02/12; BR group). We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691), NIH grants R01 NS40752 to DHG, R01NS082094 to BLF, and R01 HL130996 to WBD, the Bev Carrick Memorial Fund to GC (US group), and Cancer Research UK Programme Award C24110/A15394 to MOD. Funding sources had no specific roles.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Ramos, E.M., Carecchio, M., Lemos, R. et al. Primary brain calcification: an international study reporting novel variants and associated phenotypes. Eur J Hum Genet 26, 1462–1477 (2018). https://doi.org/10.1038/s41431-018-0185-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0185-4
This article is cited by
-
Upstream open reading frame-introducing variants in patients with primary familial brain calcification
European Journal of Human Genetics (2024)
-
Homozygous SLC20A2 mutations cause congenital CMV infection-like phenotype
Acta Neurologica Belgica (2023)
-
Expanding the genetic spectrum of primary familial brain calcification due to SLC2OA2 mutations: a case series
neurogenetics (2021)
-
MiR-9-5p Regulates Genes Linked to Cerebral Calcification in the Osteogenic Differentiation Model and Induces Generalized Alteration in the Ion Channels
Journal of Molecular Neuroscience (2021)
-
A novel PDGFRB sequence variant in a family with a mild form of primary familial brain calcification: a case report and a review of the literature
BMC Neurology (2019)