Abstract
Neural tube defects (NTDs) remain one of the most serious birth defects, and although genes in several pathways have been implicated as risk factors for neural tube defects via knockout mouse models, very few molecular causes in humans have been identified. Whole exome sequencing identified deleterious variants in key apoptotic genes in two families with recurrent neural tube defects. Functional studies in fibroblasts indicate that these variants are loss-of-function, as apoptosis is significantly reduced. This is the first report of variants in apoptotic genes contributing to neural tube defect risk in humans.
Similar content being viewed by others
Introduction
As a group, neural tube defects (NTDs) including spina bifida, anencephaly, encephalocele, and craniorachischisis occur with an incidence of approximately 1:1000 births in the United States, 78% of which are isolated defects without malformations in other systems and 22% are associated with other anomalies, in some cases constituting recognizable syndromes [1,2,3]. NTDs result from the failure of the neural tube to initiate or complete closure during the process of neurulation. The process of neurulation takes place in the first 3 weeks of gestation starting with the formation of the neural plate, the folding and apposition of the neural folds, and closure and zippering of the neural tube. The process is finalized in human after closure of the cranial and caudal neuropores on approximately days 24 and 26, respectively [4].
Neurulation is a complex process involving a number of diverse cellular pathways. Folic acid metabolism has been implicated in the etiology of NTDs, and indeed, fortification of grain products with folic acid has elicited significant reductions in NTDs across the globe, with a 28% reduction of NTD occurrence in the United States and up to 81% reduction in areas with particularly high incidence rates such as Northern China and Labrador [5]. However, NTD occurrence persists, indicating that other cellular pathways may be involved [5, 6]. Sonic hedgehog (Shh), Wnt signaling, and the planar cell polarity pathway (also referred to as the noncanonical Wnt, or PCP, pathway) are important in establishing the axis of the embryo, in driving cellular differentiation in the CNS, in convergent extension of the neural plate, and have been implicated in the development of NTDs [7,8,9]. Additionally, studies in animal models indicate that the apoptotic pathway may be an important mediator of neural tube closure. Despite the implication of almost 200 genes in NTD etiology via mouse knockout models, the only NTD-associated genes to be identified in human thus far have been limited to the PCP pathway, including VANGL1, VANGL2, CELSR1, FUZ, FZD3, FZD6, DVL2, PRICKLE1, SCRIB, DACT1, and DVL3 [10, 11], the folate pathway [12], and other genes such as PAX3 [13]. The gene T (brachyury), which is a transcription factor essential in the differentiation of cells that form the notochord during gastrulation, has also been recently associated with an NTD phenotype including sacral agenesis, persistent notochord, and abnormal ossification of the vertebra that segregated in an autosomal recessive fashion in four consanguineous families [14,15,16]. Here, we present two families with recurrent folate-resistant NTDs in which whole exome sequencing (WES) identified deleterious variants in key intrinsic apoptotic pathway genes, APAF1 and CASP9. This is the first report of variants in apoptotic pathway genes contributing to NTD risk in humans.
Materials and methods
Clinical assessment
This study was approved by the Greenwood Genetic Center (GGC) and Self Regional Healthcare Institutional Review Board. Informed consent was obtained from the families for fetal autopsy, diagnostic studies, and scientific study through the South Carolina Anatomical Gift Act. Blood samples were collected from the parents in both families, and from four unaffected siblings for segregation analysis in Family 2 (Fig. 1b; Family 2, individuals II-1, II-3, II-5, and II-7). Placental tissue (FFPE) was obtained from a second affected fetus from Family 2 (Individual II-4) from the Carolinas Medical Center (Charlotte, NC, USA). DNA was isolated using the QIAamp Blood Mini Kit for fetal fibroblasts, the Autopure LS for blood samples, or using DNeasy Tissue Kit for FFPE samples (Qiagen, Valencia, CA, USA).
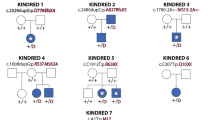
Pedigrees and clinical features of the families and affected fetuses. a Pedigree of Family 1 and b pedigree of Family 2. c (1 and 2) Clinical features of fetus II-10 in Family 2 (1, front and 2, back); 20-week gestation female fetus with craniorachischisis. c (3 and 4) Radiograph of fetus II-10 in Family 2 with multiple vertebral segmentation anomalies and ribs numbering 9 on the left side and 11 on the right. c (4) Enlarged image of the radiograph to show vertebral anomalies
Whole exome sequencing and variant analysis
Whole exome sequencing was performed as previously described [17], with the following differences: library preparation/capture was performed using the Agilent SureSelectXT Library Preparation kit and the SureSelectXT Human All Exon v5 probes (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed using the 5500 Series SOLiD XL (Life Technologies, Thermo Fisher, Carlsbad, CA, USA) for Family 1 and the Illumina NextSeq 500® Sequencing System for Family 2 (Illumina Inc., San Diego, CA, USA).
Exome analysis for Family 1 was performed using Cartagenia Bench Lab NGS (Agilent Technologies) in 2013. Briefly, the analysis was performed using population filters to identify rare variants (i.e., present with less than a 1% minor allele frequency in the public SNP databases) and phenotypic filters using Human Phenotype Ontology (HPO) terms to identify variants in genes potentially related to the phenotype. The alignment process in this proband called 27,206 variants, which were systematically categorized by creating various classification trees. The APAF1 variants were identified due to their rarity in the population, and selected as variants of interest based on publications demonstrating a role for apoptosis, and APAF1 specifically, in neural tube closure [18]. No other relevant pathogenic alterations were noted.
Exome analysis in Family 2 was performed using Cartagenia Bench Lab NGS as reported in Louie et al. [17]. Exome analysis was performed in 2015 and utilized the internally developed GGC analysis pipeline designed for review of variants for clinical exome testing. The alignment process in this proband called 53,359 variants. Initial filtering excluded variants that had a carrier frequency in the general, or ethnic-specific populations exceeding 1/30 based on the commonly referenced genomic databases (dbSNP, 1000 Genomes Phase 3, and ExAC, and ESP6500). Variants were required to have a read depth of at least 20× with a variant allele frequency of at least 22% to be considered high quality. Variants that passed the initial filters but exceeded a 1% genotype frequency in our internal database were also excluded from further consideration given the probability that most, or all, were likely recurrent technical artifacts. After filtering, the remaining variants were evaluated using HPO phenotype-based filters as well as additional manual curation. Variants identified by NGS in the APAF1 (NM_181861.1) and CASP9 genes (NM_001229.4) were confirmed via Sanger sequencing.
Cell culture
Patient and control fibroblasts, which were obtained from skin biopsies of the limbs, were grown in a 5% CO2 humidified tissue culture incubator in DMEM (Corning, Corning, NY, USA, #MT10017CV) supplemented with 15% FBS (Atlanta Biologicals, #S12450H), 1% penicillin/streptomycin (Corning, #MT30002CI), 1% non-essential amino acids (Corning, #MT25005CI), 2 mm L-glutamine (Corning, #MT25025CI), and 2 mm sodium pyruvate (Corning, #MT25000CI).
Western blots
Fibroblasts from affected fetuses (Fig. 1a and b; Family 1, individuals II-1 and II-3, Family 2, individual II-10) and controls were utilized. Western blots to detect CASP9 expression and to evaluate CASP9, caspase 3 (CASP3), and poly ADP-ribose polymerase (PARP) cleavage were performed using standard methods. Primary antibodies used to detect target proteins in 20 µg cell lysate were the following: anti-CASP9 antibody diluted 1:1000 (Cell Signaling Technology, Danvers, MA, USA, #9502), anti-PARP antibody diluted to 1:1000 (Cell Signaling Technology, #9542), and anti-CASP3 antibody diluted to 1:1000 (Cell Signaling Technology, #9668), and anti-GADPH antibody diluted to 1:10,000 (Santa Cruz Biotechnology, Inc., Dallas, TX, USA, #sc-32233). Secondary anti-mouse (Cell Signaling Technology, #7076) or anti-rabbit (Cell Signaling Technology, #7074) IgG HRP antibodies were used for detection and diluted to 1:2000. Western blots were analyzed using NIH ImageJ software as previously described [19].
Cleaved CASP9, CASP3, and PARP assays
Cells were incubated in treatment media (DMEM, 10% FBS, and 1% penicillin/streptomycin) ± 1 µm staurosporine (Cell Signaling Technology) for 8 h followed by western blotting.
TUNEL analysis
Fibroblasts were incubated in media ± 1 µM staurosporine for 8 h and stained with the TUNEL Andy Fluor 594 Apoptosis assay (Genecopia, Rockville, MD, USA) and the NucBlue Fixed Cell Stain ReadyProbes per the manufacturer’s protocol (ThermoFisher). For each assay point, images were captured with the Zeiss AX10 ObserverA1 fluorescent inverted microscope with Zen2Pro software (Zeiss, Jena, Germany) from four fields and assessed for TUNEL-positive nuclei and DAPI staining. Data were quantified as described in Zhang et al. [20].
Results
Clinical reports
Family 1
The mother was Caucasian with type A+ blood, and she was a cystic fibrosis carrier. At the age of 25 she became pregnant for the first time. The father, also Caucasian, was 26 years old, and his mother had a history of several first trimester miscarriages. Neither parent had a family history of intellectual disabilities, learning disorders, or birth defects. Consanguinity was denied. During the pregnancy, the mother took a prenatal vitamin with 800 mg folic acid and folic acid fortification of grain and cereal products had been completed for several years prior to this case. The pregnancy was notable for maternal cigarette smoking and Ambien usage.
The fetus (II-1, Fig. 1a) was an induced delivery at 21 weeks gestation (estimated by last menstrual period and ultrasound) due to fetal demise. Prenatal ultrasounds had shown spina bifida from T11-L5, hydrocephalus, posterior-inferior positioning of cerebellum leading to obliteration of cysterna magna, and scalloping of parietal bones. Fetal autopsy was notable for multi-organ hypoplasia (thymus, heart, liver, kidneys), narrow head with head circumference of 17.6 cm, and small feet. Radiographs showed lumbar kyphosis and malformed ribs including irregular placement and fusion and bifurcation of ribs 4–7 on the right. Chromosome analysis noted 46,XY karyotype which agreed with external examination of genitalia. The mother was enrolled in the South Carolina Birth Defects Surveillance and Prevention Program and began taking 4 mg of folic acid daily. In 2009, she gave birth to a healthy girl with the same partner. The delivery was at 39 weeks. No further medical information about this child is available at this time.
She became pregnant again in 2012 at the age of 30 with the same father as with her previous two pregnancies. She took the recommended 400 mcg of folic acid daily based on her history of prior NTD affected pregnancy. T10-S1 spina bifida was diagnosed by ultrasound. The pregnancy was otherwise complicated by a urinary tract infection which was treated with amoxicillin. Cigarette smoking, alcohol use, and drug use were denied. No teratogen exposure was reported. Delivery of the 20-week gestation fetus (II-3, Fig. 1a) was induced (estimated by last menstrual period) due to fetal demise. External examination noted a head circumference of 18.6 cm, small ears, depressed nasal tip, full philtrum, lumbar spina bifida, left equinovarus deformity, and bilateral rocker bottom feet. Internal examination showed shallow fissure of the left lung and pulmonary isomerism of the right lung. Multiple abnormalities of the dorsal intersegmental arteries were seen bilaterally with vascular injection. Radiographs showed 12 ribs on the right and 13 ribs on the left with bifurcation of the eighth rib. Chromosome analysis noted 46,XY karyotype which agreed with external examination of genitalia, and microarray was normal in both fetuses.
Family 2
The mother was 27 years old when she was referred for an abnormal ultrasound indicating a 20-week female fetus with craniorachischisis. Prior pregnancies, two of which were twin pregnancies, resulted in four spontaneous abortions, four liveborn infants, and one infant with anencephaly. She took 400 mcg folic acid daily for pregnancies II-5, II-6, II-7, II-8, and II-9, and took 1.2 mg of folic acid daily for 6−8 months prior to the presenting pregnancy, II-10 (Fig. 1b), and 1 month into the pregnancy. She also took Zofran for bipolar disorder. Ultrasound at 18 weeks gestation showed anencephaly. The fetus had holoanencephaly, spinal rachischisis extending from the cervical to the sacral spine, a ventriculoseptal defect, absent brachiocephalic artery, origination of the right subclavian artery from the descending aorta, rocker bottom feet, and indeterminate genitalia. Multiple vertebral segmentation anomalies were present in the cervical and thoracic spine. The ribs were thin, and numbered 9 on the left side and 11 on the right. A prior pregnancy (II-4, Fig. 1b), the mother’s third, was found to have anencephaly, multiple vertebral anomalies, and abnormal chest configuration by ultrasound at 18 weeks gestation. The mother took no prenatal vitamins or folic acid but took Zofran for bipolar disorder. Details of the fetal examination after induced delivery were not available, except that the umbilical cord had only two vessels. Chromosomal karyotype and microarray analysis were both normal. Formalin-fixed placental tissue was available for DNA extraction.
Whole exome sequencing, western blot, and TUNEL assays
Whole exome sequencing in Family 1 identified two variants, in trans, in the APAF1 (NM_181861.1) gene in both affected fetuses, c.[781C > T];[1350 C > G] (p.[(L261F)];[(C450W)]) (Fig. 1a, II-1 and II-3). The c.781C > T, p.(L261F), variant was paternally inherited and c.1350C > G, p.(C450W), was maternally inherited. WES in Family 2 revealed two variants, in trans, in the CASP9 (NM_001229.4) gene in both affected fetuses, c.[710A > C];[924dupT] (p.[(H237P)];[p.(Gly309Trpfs*3)]) (Fig. 1b, II-4 and II-10). The p.(H237P) (c.710A > C) variant was paternally inherited and a frameshift variant, c.924dupT (p.(Gly309Trpfs*3)), was maternally inherited. Segregation analysis in Family 2 revealed that three unaffected siblings inherited the maternal c.924dupT variant (Fig. 1b; individuals II-3, II-5, and II-7) and one maternal half-sibling did not inherit the c.924dupT variant (Fig. 1b, II-1). These variants have been submitted to the Leiden Open Variant Database [21] (LOVD; www.lovd.nl) and can be found associated with the following LOVD Individual ID numbers: #00108621 for the APAF1 proband (https://databases.lovd.nl/shared/genes/APAF1) and #00108623 for the CASP9 proband (https://databases.lovd.nl/shared/genes/CASP9).
In silico analyses of the APAF1 and CASP9 missense variants identified in these two families using algorithms that predict folding free energy changes induced by the variant amino acid residues (mCSM, SDM, DUET, I-mutant, PoPMuSiC, and SAFFEC) suggested they have destabilizing effects and may lead to more rapid protein degradation (Fig. 2 and Supplementary Figure 1). Analysis using web-based algorithms to predict the pathogenicity of the missense changes (PolyPhen-2, SIFT, and MutationTaster) unanimously predicted the p.(L261F) and p.(H237P) missense changes to be potentially damaging, but were discordant for the p.(C450W) variant (Fig. 2a, b). Western blot analysis of protein isolated from fibroblasts of fetus II-10 in Family 2 failed to detect any CASP9 and showed reduced CASP9 in fetuses II-1 and II-3 from Family 1 (Fig. 3a and Supplementary Figure 2). Western blots of protein from fetal fibroblasts of Family 1 did not show a significant reduction in APAF1 protein (data not shown). Western blot analyses of cleaved CASP3, CASP9, and PARP, which are markers of apoptosis, showed significant downregulation in fibroblasts of both affected fetuses from Family 1 (Fig. 3b and Supplementary Figure 3). These activated apoptotic components were completely absent in the fibroblasts of fetus II-10 from Family 2 (Fig. 3b and Supplementary Figure 3). TUNEL assays also showed significantly reduced DNA cleavage in fibroblasts from all three affected fetuses compared to control (Fig. 3c and Supplementary Figure 4).
In silico predictions of folding free energy changes in kcal/mol (∆∆G < 0 indicates the variant destabilizes the protein while ∆∆G > 0 indicates stabilization) and predicted deleteriousness of missense variants in the APAF1 (a) and CASP9 (b) proteins. Structural modeling of the APAF1 monomer (c) and heptamer (d) with the location of the missense variants from Family 1 indicated in red. Mutation Cutoff Scanning Matrix (mCSM) [33], Site Directed Mutator (SDM) [34], DUET [35], I-Mutant2.0 [36], PoPMuSiC [37], and Single Amino Acid Folding free Energy Changes (SAAFEC) [38] were used to predict folding free energy changes of missense variants identified in this study and PolyPhen-2 [39], Sort Intolerant From Tolerant (SIFT) [40], and MutationTaster2 [41] (a and b). Structural modeling in c and d was performed using the PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC
Functional studies examining expression of CASP9 and downstream apoptotic pathway activation. a Quantification of western blot of CASP9 in control and affected fetuses II-1 and II-3 in Family 1 and affected fetus II-10 in Family 2. Level of CASP9 protein is significantly reduced in all fetal fibroblast cells and almost absent in Family 2 fetus II-10 with the CASP9 variants in trans (*p < 0.05, **p < 0.01, ***p < 0.001). b Quantification of activation of downstream apoptotic factors. Level of cleaved CASP9, CASP3, and PARP was significantly reduced in fibroblast cells from all three fetuses as compared with control (*p < 0.01, **p < 0.001, ***p < 0.0001). c TUNEL assay in control and fetal fibroblasts. Significant reduction in apoptosis was observed in NTD fetus cell lines compared to control (*p < 0.01, **p < 0.001). Student’s t-test was used to compare data from patient and control fibroblasts. Graphs represent the averages and standard deviation for experiments performed in triplicate
Discussion
Apoptosis has been studied extensively in association with neural tube closure; both decreased as well as increased apoptotic activity have been shown to lead to NTDs in mice [18, 22,23,24,25,26]. The variants identified in this study were compound heterozygous loss-of-function changes, in trans, in APAF1 and CASP9 in families with recurrent folate-resistant NTDs.
The APAF1 variants identified in Family 1, p.[(L261F)];[(C450W)], (c.[781C > T];[1350C > G]), are rare in the population databases (minor allele frequencies (MAF) of 0.012% and 0.018% in the Exome Aggregation Consortium database, and of 0.0267 and 0.0167% in the gnomAD browser, respectively). The p.L261F variant is located in an NOD/NBARC functional domain (www.uniprot.com), and both missense variants are located in the “hub” of the assembled apoptosome (Fig. 2c, d). The NOD/NBARC domain is predicted to function in ATP or ADP hydrolysis, which is critical for the conformational change required to activate APAF1 [27, 28]. Functional studies of fibroblasts from the fetuses in Family 1 demonstrated that, while the two variants do not appear to destabilize the APAF1 protein as predicted by the in silico algorithms that predicted a change in folding free-energy due to these variants (Fig. 2a), the apoptotic activity in these cells was deficient compared to controls (Fig. 3c and Supplementary Figure 4). It is possible that the overall predicted change in the folding free energy of about 1 kcal/mol for these missense variants is insufficient to cause the APAF1 protein to unfold, and while structure-based predictors provide information about potential molecular mechanisms of disease, there may be cases in which disease is caused by factors other than protein stability.
The CASP9 p.[(H237P)];[p.(Gly309Trpfs*3)], (c.[710A > C];[924dupT]), variants identified in Family 2 are rare in the population (MAF of 0.15 and 0.00% in ExAC, and 0.14 and 0.0046% in gnomAD, respectively) and in silico algorithms predict p.(H237P) to be deleterious (Fig. 2b). The p.(H237P) variant disrupts one of two catalytic residues that are highly conserved in caspases 1, 3, 7, 8, and 9 [29]. The p.(H237P) variant has also been reported in HGMD in individuals with immunodeficiency, and functional studies demonstrated that the p.(H237P) protein failed to associate with APAF1 to form the mature apoptosome in vitro [30]. Functional studies in the fibroblasts of fetus II-10 in Family 2 demonstrated that not only was the CASP9 protein destabilized in these cells, consistent with the in silico predictions (Fig. 2b), it was completely absent (Fig. 3a). Additionally, the apoptotic activity in these cells, as measured by TUNEL staining, was reduced (Fig. 3c), and cleaved CASP3 and PARP were not detectable (Fig. 3b).
Mouse model studies have indicated that apoptotic gene products are important for neurulation. Casp9 and Apaf1 knockout mice, as well as mice with Apaf1 deficiency, exhibit exencephaly [18, 22, 23, 31]. Although the exact mechanism is not yet clear, these results indicate that Casp9 and Apaf1 are key players in neural tube closure in mice. One recent study concluded that apoptosis is not necessary for neural tube closure based on temporary chemical inhibition of apoptosis at the onset of neurulation. However, even that study demonstrated that if Apaf1 and Casp3 are knocked out genetically, neural tube closure is defective and results in exencephaly [32]. These results indicate that the genetic deficiency of these factors over the course of development (as with loss-of-function variants or gene deletion), from fertilization through development, is more deleterious to embryonic neurulation than temporal apoptotic deficiency. The developmental effects of complete loss of apoptotic genes is expanded upon by Yamaguchi et al. [24] wherein live imaging of neurulation in Apaf1 and Casp3 knockout mice showed reductions in the bending of the neural plate and slowing or stalling of neural tube closure. Results showed that one-quarter of the Apaf1-/- mice and one-half of the Casp3-/- mice exhibited exencephaly as a result of this slowing or stalling [24]. Although the exact role of apoptosis in development and neurulation requires further elucidation, it is clear from mouse studies that apoptosis plays an integral part in these processes.
In summary, this is the first report demonstrating that deleterious variants in the apoptotic genes APAF1 and CASP9, in human fetuses, recapitulate the phenotype observed in knockout mouse models. Taken together, the identification of compound heterozygous loss-of-function variants in key apoptotic genes in fetuses with NTDs presented here, along with previous studies demonstrating that knockout of apoptotic genes is associated with NTDs in mouse, suggest that the loss of key apoptotic gene products is a significant risk factor for NTDs.
Data availability
The data sets analyzed during the current study are available from the corresponding author upon reasonable request.
References
Mitchell LE. Epidemiology of neural tube defects. Am J Med Genet C Semin Med Genet 2005;135C:88–94.
Stevenson RE, Allen WP, Pai GS, et al. Decline in prevalence of neural tube defects in a high-risk region of the United States. Pediatrics 2000;106:677–83.
Bupp CP, Sarasua SM, Dean JH, Stevenson RE. When folic acid fails: insights from 20 years of neural tube defect surveillance in South Carolina. Am J Med Genet A 2015;167A:2244–50.
Sadler TW. Embryology of neural tube development. Am J Med Genet C Semin Med Genet 2005;135C:2–8.
Heseker H. Folic acid and other potential measures in the prevention of neural tube defects. Ann Nutr Metab 2011;59:41–5.
Heseker HB, Mason JB, Selhub J, Rosenberg IH, Jacques PF. Not all cases of neural-tube defect can be prevented by increasing the intake of folic acid. Br J Nutr 2009;102:173–80.
Murdoch JN, Copp AJ. The relationship between Sonic hedgehog signalling, cilia and neural tube defects. Birt Defects Res A Clin Mol Teratol 2010;88:633–52.
Yu W, McDonnell K, Taketo MM, Bai CB. Wnt signaling determines ventral spinal cord cell fates in a time-dependent manner. Development 2008;135:3687–96.
Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis 2008;4:68–75.
Harris MJ, Juriloff DM. Mouse mutants with neural tube closure defects and their role in understanding human neural tube defects. Birt Defects Res A Clin Mol Teratol 2007;79:187–210.
Cai C, Shi O. Genetic evidence in planar cell polarity signaling pathway in human neural tube defects. Front Med 2014;8:68–78.
Imbard A, Benoist J-F, Blom HJ. Neural tube defects, folic acid and methylation. Int J Environ Res Public Health 2013;10:4352–89.
Milunsky JM. Waardenburg Syndrome Type I. In: Pagon RA, Adam MP, Ardinger HH et al., editors. GeneReviews(®). Seattle, WA: University of Washington, Seattle; 1993. http://www.ncbi.nlm.nih.gov/books/NBK1531/. Accessed 23 May 2017.
Postma AV, Alders M, Sylva M, et al. Mutations in the T (brachyury) gene cause a novel syndrome consisting of sacral agenesis, abnormal ossification of the vertebral bodies and a persistent notochordal canal. J Med Genet 2014;51:90–97.
Shaheen R, Alshail E, Alaqeel A, Ansari S, Hindieh F, Alkuraya FS, T (brachyury) is linked to a Mendelian form of neural tube defects in humans. Hum Genet. 2015;134:1139–41.
Zhu J, Kwan KM, Mackem S. Putative oncogene Brachyury (T) is essential to specify cell fate but dispensable for notochord progenitor proliferation and EMT. Proc Natl Acad Sci USA 2016;113:3820–25.
Louie RJ, Tan QK-G, Gilner JB, et al. Novel pathogenic variants in FOXP3 in fetuses with echogenic bowel and skin desquamation identified by ultrasound. Am J Med Genet A 2017;173:1219–25.
Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell 1998;94:727–37.
Zhang Z, Norris J, Kalscheuer V, et al. A Y328C missense mutation in spermine synthase causes a mild form of Snyder−Robinson syndrome. Hum Mol Genet 2013;22:3789–97.
Zhang W, Shi HY, Zhang M. Maspin overexpression modulates tumor cell apoptosis through the regulation of Bcl-2 family proteins. BMC Cancer 2005;5:50.
Fokkema IFAC, Taschner PEM, Schaafsma GCP, Celli J, Laros JFJ, den Dunnen JT, LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–63.
Hakem R, Hakem A, Duncan GS, et al. Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell. 1998;94:339–52
Kuida K, Haydar TF, Kuan CY, et al. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–37.
Yamaguchi Y, Shinotsuka N, Nonomura K, et al. Live imaging of apoptosis in a novel transgenic mouse highlights its role in neural tube closure. J Cell Biol. 2011;195:1047–60.
Homanics GE, Maeda N, Traber MG, Kayden HJ, Dehart DB, Sulik KK. Exencephaly and hydrocephaly in mice with targeted modification of the apolipoprotein B (Apob) gene. Teratology 1995;51:1–10.
Ikeda A, Ikeda S, Gridley T, Nishina PM, Naggert JK. Neural tube defects and neuroepithelial cell death in Tulp3 knockout mice. Hum Mol Genet 2001;10:1325–34.
van der Biezen EA, Jones JDG. The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol 1998;8:R226–R228.
Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. J Cell Sci 2010;123:3209–14.
Denault J-B, Salvesen GS. Caspases: keys in the ignition of cell death. Chem Rev 2002;102:4489–4500.
Clemente N, Boggio E, Gigliotti CL, et al. A mutation in caspase-9 decreases the expression of BAFFR and ICOS in patients with immunodeficiency and lymphoproliferation. Genes Immun 2015;16:151–61.
Honarpour N, Gilbert SL, Lahn BT, Wang X, Herz J. Apaf-1 deficiency and neural tube closure defects are found in fog mice. Proc Natl Acad Sci USA 2001;98:9683–87.
Massa V, Savery D, Ybot-Gonzalez P, et al. Apoptosis is not required for mammalian neural tube closure. Proc Natl Acad Sci USA 2009;106:8233–38.
Pires DEV, Ascher DB, Blundell TL. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014;30:335–42.
Worth CL, Preissner R, Blundell TL. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res 2011;39:W215–W222.
Pires DEV, Ascher DB, Blundell TL. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res 2014;42:W314–W319.
Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 2005;33:W306–W310.
Gilis D, Rooman M. PoPMuSiC, an algorithm for predicting protein mutant stability changes. Application to prion proteins. Protein Eng Des Sel 2000;13:849–56.
Getov I, Petukh M, Alexov E. SAAFEC: predicting the effect of single point mutations on protein folding free energy using a knowledge-modified MM/PBSA approach. Int J Mol Sci 2016;17:512.
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–49.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073–81.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014;11:361–62.
Acknowledgements
Funding for this work included grants from the South Carolina Department of Health and Environmental Control (SCDHEC), the South Carolina Department of Disabilities and Special Needs (SCDDSN), the National Institute of General Medical Sciences (NIH R01Gm93937, NIGMS), and the GGC Foundation. We would also like to thank the patients and their families for their participation in this study and recognize the contributions of our laboratory staff and the clinical team at the Greenwood Genetic Center for their considerable efforts.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Spellicy, C., Norris, J., Bend, R. et al. Key apoptotic genes APAF1 and CASP9 implicated in recurrent folate-resistant neural tube defects. Eur J Hum Genet 26, 420–427 (2018). https://doi.org/10.1038/s41431-017-0025-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-017-0025-y
This article is cited by
-
SNIP1 and PRC2 coordinate cell fates of neural progenitors during brain development
Nature Communications (2023)
-
Apoptotic cell death in disease—Current understanding of the NCCD 2023
Cell Death & Differentiation (2023)
-
Genetic diagnosis in first or second trimester pregnancy loss using exome sequencing: a systematic review of human essential genes
Journal of Assisted Reproduction and Genetics (2019)