Abstract
Background/Objectives
Curcumin, a natural polyphenol compound in the spice turmeric, has been found to have potent anti-oxidative and anti-inflammatory activity. Curcumin may treat non-alcoholic fatty liver disease (NAFLD) through its beneficial effects on biomarkers of oxidative stress (OS) and inflammation, which are considered as two feature of this disease. However, the effects of curcumin on NAFLD have been remained poorly understood. This investigation evaluated the effects of administrating curcumin on metabolic status in NAFLD patients.
Subjects/Methods
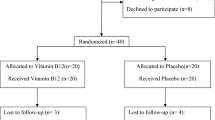
Fifty-eight NAFLD patients participated in a randomized, double-blind, placebo-controlled parallel design of study. The subjects were allocated randomly into two groups, which either received 250 mg phospholipid curcumin or placebo, one capsule per day for a period of 8 weeks. Fasting blood samples were taken from each subject at the start and end of the study period. Subsequently, metabolomics analysis was performed for serum samples using NMR.
Results
Compared with the placebo, supplementing phospholipid curcumin resulted in significant decreases in serum including 3- methyl-2-oxovaleric acid, 3-hydroxyisobutyrate, kynurenine, succinate, citrate, α-ketoglutarate, methylamine, trimethylamine, hippurate, indoxyl sulfate, chenodeoxycholic acid, taurocholic acid, and lithocholic acid. This profile of metabolic biomarkers could distinguish effectively NAFLD subjects who were treated with curcumin and placebo groups, achieving value of 0.99 for an area under receiver operating characteristic curve (AUC).
Conclusions
Characterizing the serum metabolic profile of the patients with NAFLD at the end of the intervention using NMR-based metabolomics method indicated that the targets of curcumin treatment included some amino acids, TCA cycle, bile acids, and gut microbiota.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313:2263–73.
Bacon BR, Farahvash MJ, Janney CG, Neuschwander-Tetri BA. Nonalcoholic steatohepatitis: an expanded clinical entity. Gastroenterology. 1994;107:1103–9.
Matteoni CA, Younossi ZM, Gramlich T, Boparai N, Liu YC, McCullough AJ. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116:1413–9.
Lindor KD, Kowdley KV, Heathcote EJ, Harrison ME, Jorgensen R, Angulo P, et al. Ursodeoxycholic acid for treatment of nonalcoholic steatohepatitis: results of a randomized trial. Hepatology. 2004;39:770–8.
Lavine JE, Schwimmer JB, Van Natta ML, Molleston JP, Murray KF, Rosenthal P, et al. Effect of vitamin E or metformin for treatment of nonalcoholic fatty liver disease in children and adolescents: the TONIC randomized controlled trial. JAMA. 2011;305:1659–68.
Aggarwal BB, Sundaram C, Malani N, Ichikawa H. Curcumin: the Indian solid gold. The molecular targets and therapeutic uses of curcumin in health and disease. Springer, New York, 2007. p. 1–75.
Aggarwal BB, Kumar A, Bharti AC. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res. 2003;23(1/A):363–98.
Babu PS, Srinivasan K. Hypolipidemic action of curcumin, the active principle of turmeric (Curcuma longa) in streptozotocin induced diabetic rats. Mol Cell Biochem. 1997;166:169–75.
Jang E-M, Choi M-S, Jung UJ, Kim M-J, Kim H-J, Jeon S-M, et al. Beneficial effects of curcumin on hyperlipidemia and insulin resistance in high-fat–fed hamsters. Metabolism. 2008;57:1576–83.
Jurenka JS. Anti-inflammatory properties of curcumin, a major constituent of Curcuma longa: a review of preclinical and clinical research. Altern Med Rev. 2009;14:141–53.
Manjunatha H, Srinivasan K. Hypolipidemic and antioxidant effects of curcumin and capsaicin in high-fat-fed rats. Can J Physiol Pharmacol. 2007;85:588–96.
Schraufstatter E, Bernt H. Antibacterial action of curcumin and related compounds. Nature. 1949;164:456–7.
Nishimura Y, Kitagishi Y, Yoshida H, Okumura N, Matsuda S. Ethanol extracts of black pepper or turmeric down-regulated SIRT1 protein expression in Daudi culture cells. Mol Med Rep. 2011;4:727–30.
Elahi RK. Preventive effects of turmeric (Curcuma longa Linn.) powder on hepatic steatosis in the rats fed with high fat diet. Life Sci. 2012;9:5462–8.
Raftery D. High-throughput NMR spectroscopy. Anal Bioanal Chem. 2004;378:1403–4.
Lacey ME, Subramanian R, Olson DL, Webb AG, Sweedler JV. High-resolution NMR spectroscopy of sample volumes from 1 nL to 10 μL. Chem Rev. 1999;99:3133–52.
Nobakht M, Gh BF, Aliannejad R, Rezaei-Tavirani M, Taheri S, Oskouie AA. The metabolomics of airway diseases, including COPD, asthma and cystic fibrosis. Biomarkers. 2015;20:5–16.
Emwas A-HM, Salek RM, Griffin JL, Merzaban J. NMR-based metabolomics in human disease diagnosis: applications, limitations, and recommendations. Metabolomics. 2013;9:1048–72.
Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–83.
Stadlmayr A, Aigner E, Steger B, Scharinger L, Lederer D, Mayr A, et al. Nonalcoholic fatty liver disease: an independent risk factor for colorectal neoplasia. J Intern Med. 2011;270:41–9.
Feldman A, Eder SK, Felder TK, Kedenko L, Paulweber B, Stadlmayr A, et al. Clinical and metabolic characterization of lean Caucasian subjects with non-alcoholic fatty liver. Am J Gastroenterol. 2017;112:102–10.
Yin P, Lehmann R, Xu G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal Bioanal Chem. 2015;407:4879–92.
Nobakht BF, Arefi Oskouie A, Rezaei-Tavirani M, Aliannejad R, Taheri S, Fathi F, et al. NMR spectroscopy-based metabolomic study of serum in sulfur mustard exposed patients with lung disease. Biomarkers. 2017;22:413–9.
Nobakht BF, Aliannejad R, Rezaei-Tavirani M, Arefi Oskouie A, Naseri MT, Parastar H, et al. NMR-and GC/MS-based metabolomics of sulfur mustard exposed individuals: a pilot study. Biomarkers. 2016;21:479–89.
Brown FF, Campbell ID, Kuchel PW, Rabenstein DC. Human erythrocyte metabolism studies by 1 H spin echo NMR. FEBS Lett. 1977;82:12–6.
Tang H, Wang Y, Nicholson JK, Lindon JC. Use of relaxation-edited one-dimensional and two dimensional nuclear magnetic resonance spectroscopy to improve detection of small metabolites in blood plasma. Anal Biochem. 2004;325:260–72.
Fardet A, Canlet C, Gottardi G, Lyan B, Llorach R, Rémésy C, et al. Whole-grain and refined wheat flours show distinct metabolic profiles in rats as assessed by a 1H NMR-based metabonomic approach. J Nutr. 2007;137:923–9.
Viant MR. Improved methods for the acquisition and interpretation of NMR metabolomic data. Biochem Biophys Res Commun. 2003;310:943–8.
Mahadevan S, Shah SL, Marrie TJ, Slupsky CM. Analysis of metabolomic data using support vector machines. Anal Chem. 2008;80:7562–70.
Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67:793–811.
Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, et al. The human serum metabolome. PLoS ONE. 2011;6:e16957.
Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–60.
Xia J, Broadhurst DI, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9:280–99.
Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–W7.
Booth SC, Weljie AM, Turner RJ. Computational tools for the secondary analysis of metabolomics experiments. Comput Struct Biotechnol J. 2013;4:e201301003.
Shao W, Yu Z, Chiang Y, Yang Y, Chai T, Foltz W, et al. Curcumin prevents high fat diet induced insulin resistance and obesity via attenuating lipogenesis in liver and inflammatory pathway in adipocytes. PLoS ONE. 2012;7:e28784.
El-Moselhy MA, Taye A, Sharkawi SS, El-Sisi SF, Ahmed AF. The antihyperglycemic effect of curcumin in high fat diet fed rats. Role of TNF-α and free fatty acids. Food Chem Toxicol. 2011;49:1129–40.
Abbondante S, Eckel-Mahan KL, Ceglia NJ, Baldi P, Sassone-Corsi P. Comparative circadian metabolomics reveal differential effects of nutritional challenge in the serum and liver. J Biol Chem. 2016;291:2812–28.
Soga T, Sugimoto M, Honma M, Mori M, Igarashi K, Kashikura K, et al. Serum metabolomics reveals γ-glutamyl dipeptides as biomarkers for discrimination among different forms of liver disease. J Hepatol. 2011;55:896–905.
Denery JR, Nunes AA, Hixon MS, Dickerson TJ, Janda KD. Metabolomics-based discovery of diagnostic biomarkers for onchocerciasis. PLoS Negl Trop Dis. 2010;4:e834.
Tan Y, Yin P, Tang L, Xing W, Huang Q, Cao D, et al. Metabolomics study of stepwise hepatocarcinogenesis from the model rats to patients: potential biomarkers effective for small hepatocellular carcinoma diagnosis. Mol Cell Proteom. 2012;11:M111. 010694
Hori S, Nishiumi S, Kobayashi K, Shinohara M, Hatakeyama Y, Kotani Y, et al. A metabolomic approach to lung cancer. Lung Cancer. 2011;74:284–92.
Xuan J, Pan G, Qiu Y, Yang L, Su M, Liu Y, et al. Metabolomic profiling to identify potential serum biomarkers for schizophrenia and risperidone action. J Proteome Res. 2011;10:5433–43.
Townsend MK, Bao Y, Poole EM, Bertrand KA, Kraft P, Wolpin BM, et al. Impact of pre-analytic blood sample collection factors on metabolomics. Cancer Epidemiol Biomark Prev. 2016;25:823–29.
Brauer R, Leichtle AB, Fiedler GM, Thiery J, Ceglarek U. Preanalytical standardization of amino acid and acylcarnitine metabolite profiling in human blood using tandem mass spectrometry. Metabolomics. 2011;7:344–52.
Ishikawa S, Sugimoto M, Kitabatake K, Tu M, Sugano A, Yamamori I, et al. Effect of timing of collection of salivary metabolomic biomarkers on oral cancer detection. Amino Acids. 2017;49:761–70.
Emwas A-H, Roy R, McKay RT, Ryan D, Brennan L, Tenori L, et al. Recommendations and standardization of biomarker quantification using NMR-based metabolomics with particular focus on urinary analysis. J Proteome Res. 2016;15:360–73.
Rubio-Aliaga I, de Roos B, Duthie SJ, Crosley LK, Mayer C, Horgan G, et al. Metabolomics of prolonged fasting in humans reveals new catabolic markers. Metabolomics. 2011;7:375–87.
Munro H, Fernstrom J, Wurtman R. Insulin, plasma aminoacid imbalance, and hepatic coma. Lancet. 1975;305:722–4.
Cheng S, Wiklund P, Autio R, Borra R, Ojanen X, Xu L, et al. Adipose tissue dysfunction and altered systemic amino acid metabolism are associated with non-alcoholic fatty liver disease. PLoS ONE. 2015;10:e0138889.
Lai Y-S, Chen W-C, Kuo T-C, Ho C-T, Kuo C-H, Tseng YJ, et al. Mass-spectrometry-based serum metabolomics of a C57BL/6J mouse model of high-fat-diet-induced non-alcoholic fatty liver disease development. J Agric Food Chem. 2015;63:7873–84.
Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–26.
Lake AD, Novak P, Shipkova P, Aranibar N, Robertson DG, Reily MD, et al. Branched chain amino acid metabolism profiles in progressive human nonalcoholic fatty liver disease. Amino Acids. 2015;47:603–15.
King NJ, Thomas SR. Molecules in focus: indoleamine 2, 3-dioxygenase. Int J Biochem Cell Biol. 2007;39:2167–72.
O’Connor JC, André C, Wang Y, Lawson MA, Szegedi SS, Lestage J, et al. Interferon-γ and tumor necrosis factor-α mediate the upregulation of indoleamine 2, 3-dioxygenase and the induction of depressive-like behavior in mice in response to bacillus Calmette-Guérin. J Neurosci. 2009;29:4200–9.
Connor TJ, Starr N, O’Sullivan JB, Harkin A. Induction of indolamine 2, 3-dioxygenase and kynurenine 3-monooxygenase in rat brain following a systemic inflammatory challenge: a role for IFN-γ? Neurosci Lett. 2008;441:29–34.
Pawlak K, Domaniewski T, Mysliwiec M, Pawlak D. The kynurenines are associated with oxidative stress, inflammation and the prevalence of cardiovascular disease in patients with end-stage renal disease. Atherosclerosis. 2009;204:309–14.
Jeong Y-I, Kim SW, Jung ID, Lee JS, Chang JH, Lee C-M, et al. Curcumin suppresses the induction of indoleamine 2, 3-dioxygenase by blocking the Janus-activated kinase-protein kinase Cδ-STAT1 signaling pathway in interferon-γ-stimulated murine dendritic cells. J Biol Chem. 2009;284:3700–8.
Jakobsdottir G, Xu J, Molin G, Ahrne S, Nyman M. High-fat diet reduces the formation of butyrate, but increases succinate, inflammation, liver fat and cholesterol in rats, while dietary fibre counteracts these effects. PLoS ONE. 2013;8:e80476.
Aragonès G, Auguet T, Berlanga A, Guiu-Jurado E, Martinez S, Armengol S, et al. Increased circulating levels of alpha-ketoglutarate in morbidly obese women with non-alcoholic fatty liver disease. PLoS ONE. 2016;11:e0154601.
van de Wier B, Balk JM, Haenen GR, Giamouridis D, Bakker JA, Bast BC, et al. Elevated citrate levels in non‐alcoholic fatty liver disease: the potential of citrate to promote radical production. FEBS Lett. 2013;587:2461–6.
Satapati S, Kucejova B, Duarte JA, Fletcher JA, Reynolds L, Sunny NE, et al. Mitochondrial metabolism mediates oxidative stress and inflammation in fatty liver. J Clin Invest. 2015;125:4447–62.
Sunny NE, Parks EJ, Browning JD, Burgess SC. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metab. 2011;14:804–10.
Begriche K, Massart J, Robin MA, Bonnet F, Fromenty B. Mitochondrial adaptations and dysfunctions in nonalcoholic fatty liver disease. Hepatology. 2013;58:1497–507.
Pérez‐Carreras M, Del Hoyo P, Martín MA, Rubio JC, Martín A, Castellano G, et al. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology. 2003;38:999–1007.
García-Ruiz I, Fernández-Moreira D, Solís-Muñoz P, Rodríguez-Juan C, Díaz-Sanjuán T, Muñoz-Yagüe T, et al. Mitochondrial complex I subunits are decreased in murine nonalcoholic fatty liver disease: implication of peroxynitrite. J Proteome Res. 2010;9:2450–9.
Gusdon AM, Song K-X, Qu S. Nonalcoholic fatty liver disease: pathogenesis and therapeutics from a mitochondria-centric perspective. Oxid Med Cell Longev. 2014;2014:637027.
Jung KH, Park J-W. Suppression of mitochondrial NADP + -dependent isocitrate dehydrogenase activity enhances curcumin-induced apoptosis in HCT116 cells. Free Radic Res. 2011;45:431–8.
Shah F-A, Gim S-A, Sung J-H, Jeon S-J, Kim M-O, Koh P-O. Identification of proteins regulated by curcumin in cerebral ischemia. J Surg Res. 2016;201:141–8.
Rastogi M, Ojha RP, Rajamanickam G, Agrawal A, Aggarwal A, Dubey G. Curcuminoids modulates oxidative damage and mitochondrial dysfunction in diabetic rat brain. Free Radic Res. 2008;42:999–1005.
Boursier J, Diehl AM. Implication of gut microbiota in nonalcoholic fatty liver disease. PLoS Pathog. 2015;11:e1004559.
Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–24.
Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85.
Targher G, Byrne CD. Non-alcoholic fatty liver disease: an emerging driving force in chronic kidney disease. Nat Rev Nephrol. 2017;13:297–310.
Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–703.
Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–81.
Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23:859–68.
Shen L, Liu L, Ji H-F. Regulative effects of curcumin spice administration on gut microbiota and its pharmacological implications. Food Nutr Res. 2017;61:1361780.
Feng W, Wang H, Zhang P, Gao C, Tao J, Ge Z, et al. Modulation of gut microbiota contributes to curcumin-mediated attenuation of hepatic steatosis in rats. Biochim Biophys Acta. 2017;1861:1801–12.
Kalhan SC, Guo L, Edmison J, Dasarathy S, McCullough AJ, Hanson RW, et al. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metab Clin Exp. 2011;60:404–13.
Mouzaki M, Wang AY, Bandsma R, Comelli EM, Arendt BM, Zhang L, et al. Bile acids and dysbiosis in non-alcoholic fatty liver disease. PLoS ONE. 2016;11:e0151829.
Ferslew BC, Xie G, Johnston CK, Su M, Stewart PW, Jia W, et al. Altered bile acid metabolome in patients with nonalcoholic steatohepatitis. Dig Dis Sci. 2015;60:3318–28.
Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol. 2009;15:1677–89.
Yang F, Tang X, Ding L, Yang Q, Gong J, Wang G, et al. Curcumin protects ANIT-induced cholestasis through signaling pathway of FXR-regulated bile acid and inflammation. Sci Rep. 2016;6:33052.
Funding
This work was supported financially by the grant, which has been taken from Neyshabur University of Medical Science (NUMS) and Sharif University of Technology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chashmniam, S., Mirhafez, S.R., Dehabeh, M. et al. A pilot study of the effect of phospholipid curcumin on serum metabolomic profile in patients with non-alcoholic fatty liver disease: a randomized, double-blind, placebo-controlled trial. Eur J Clin Nutr 73, 1224–1235 (2019). https://doi.org/10.1038/s41430-018-0386-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41430-018-0386-5
This article is cited by
-
Modeling dry eye with an air–liquid interface in corneal epithelium-on-a-chip
Scientific Reports (2024)
-
An updated meta-analysis of effects of curcumin on metabolic dysfunction-associated fatty liver disease based on available evidence from Iran and Thailand
Scientific Reports (2023)
-
Traditional Chinese Medicine in nonalcoholic fatty liver disease: molecular insights and therapeutic perspectives
Chinese Medicine (2021)
-
Effects of resistance training and turmeric supplementation on reactive species marker stress in diabetic rats
BMC Sports Science, Medicine and Rehabilitation (2020)