Abstract
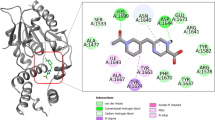
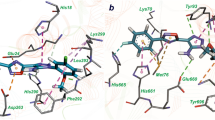
InhA or enoyl-acyl carrier protein reductase of Mycobacterium tuberculosis (mtInhA), which controls mycobacterial cell wall construction, has been targeted in the development of antituberculosis drugs. Previously, our in silico structure-based drug screening study identified a novel class of compounds (designated KES4), which is capable of inhibiting the enzymatic activity of mtInhA, as well as mycobacterial growth. The compounds are composed of four ring structures (A–D), and the MD simulation predicted specific interactions with mtInhA of the D-ring and methylene group between the B-ring and C-ring; however, there is still room for improvement in the A-ring structure. In this study, a structure–activity relationship study of the A-ring was attempted with the assistance of in silico docking simulations. In brief, the virtual chemical library of A-ring-modified KES4 was constructed and subjected to in silico docking simulation against mtInhA using the GOLD program. Among the selected candidates, we achieved synthesis of seven compounds, and the bioactivities (effects on InhA activity and mycobacterial growth and cytotoxicity) of the synthesized molecules were evaluated. Among the compounds tested, two candidates (compounds 3d and 3f) exhibited superior properties as mtInhA-targeted anti-infectives for mycobacteria than the lead compound KES4.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328:856–61.
Das P, Horton R. Tuberculosis-time to accelerate progress. Lancet. 2010;375:1755–7.
Das P, Horton R. Tuberculosis-getting to zero. Lancet. 2015;386:2231–2.
Lönnroth K, Castro KG, Chakaya JM, Chauhan LS, Floyd K, Glaziou P, et al. Tuberculosis control and elimination 2010-50: cure, care, and social development. Lancet. 2010;375:1814–29.
Sotgiu G, Centis R, D’Ambrosio L, Migliori GB. Tuberculosis treatment and drug regimens. Cold Spring Harb Perspect Med. 2015;5:a017822.
Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, et al. InhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–30.
Parikh SL, Xiao G, Tonge PJ. Inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry. 2000;39:7645–50.
McMurry LM, McDermott PF, Levy SB. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob Agents Chemother. 1999;43:711–3.
Quemard A, Sacchettini JC, Dessen A, Vilcheze C, Bittman R, Jacobs WR Jr, et al. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry. 1995;34:8235–41.
Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63.
Tonge PJ, Kisker C, Slayden RA. Development of modern InhA inhibitors to combat drug resistant strains of Mycobacterium tuberculosis. Curr Top Med Chem. 2007;7:489–98.
Rotta M, Pissinate K, Villela AD, Back DF, Timmers LF, et al. Piperazine derivatives: synthesis, inhibition of the Mycobacterium tuberculosis enoyl-acyl carrier protein reductase and SAR studies. Eur J Med Chem. 2015;90:36–447.
Vilcheze C, Morbidoni HR, Weisbrod TR, Iwamoto H, Kuo M, et al. Inactivation of the inhA-encoded fatty acid synthase II (FASII) enoyl-acyl carrier protein reductase induces accumulation of the FASI end products and cell lysis of Mycobacterium smegmatis. J Bacteriol. 2000;182:59–4067.
Oliveira JS, Pereira JH, Canduri F, Rodrigues NC, de Souza ON, de Azevedo WF Jr, et al. Crystallographic and pre-steady-state kinetics studies on binding of NADH to wild-type and isoniazid-resistant enoyl-ACP(CoA) reductase enzymes from Mycobacterium tuberculosis. J Mol Biol. 2006;359:646–66.
Taira J, Ito T, Nakatani H, Umei T, Baba H, Kawashima S, et al. In silico structure-based drug screening of novel antimycobacterial pharmacophores by DOCK-GOLD tandem screening. Int J Mycobacteriol. 2017;6:142–8.
Taira J, Morita K, Kawashima S, Umei T, Baba H, Maruoka T, et al. Identification of a novel class of small compounds with anti-tuberculosis activity by in silico structure-based drug screening. J Antibiot. 2017;70:1057–64.
Kinjo T, Koseki Y, Kobayashi M, Yamada A, Morita K, Yamaguchi K, et al. Identification of compounds with potential antibacterial activity against Mycobacterium through structure-based drug screening. J Chem Inf Model. 2013;53:1200–12.
Koseki Y, Kinjo T, Kobayashi M, Aoki S. Identification of novel antimycobacterial chemical agents through the in silico multi-conformational structure-based drug screening of a large-scale chemical library. Eur J Med Chem. 2013;60:333–39.
Kanetaka H, Koseki Y, Taira J, Umei T, Komatsu H, Sakamoto H, et al. Discovery of InhA inhibitors with anti-mycobacterial activity through a matched molecular pair approach. Eur J Med Chem. 2015;94:378–85.
He X, Alian A, Stroud R, Ortiz, de Montellano PR. Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis. J Med Chem. 2006;49:6308–23.
Kobayashi M, Kinjo T, Koseki Y, Bourne CR, Barrow WW, Aoki S. Identification of novel potential antibiotics against Staphylococcus using structure-based drug screening targeting dihydrofolate reductase. J Chem Inf Model. 2014;54:1242–53.
Koseki Y, Aoki S. Computational medicinal chemistry for rational drug design: Identification of novel chemical structures with potential anti-tuberculosis activity. Curr Top Med Chem. 2014;14:176–88.
Schmidt CW. TOX 21: new dimensions of toxicity testing. Environ Health Perspect. 2009;117:A348.
Faulon JL, Visco DP, Pophale RS. The signature molecular descriptor. 1. Using extended valence sequences in QSAR and QSPR studies. J Chem Info Comput Sci. 2003;43:707–20.
Berenger F. MolEnc: a molecular encoder using rdkit and OCaml. 2019. https://github.com/UnixJunkie/molenc. Accessed 19 Nov 2019.
Breiman L. Bagging predictors. Mach Learn. 1996;24:123–40.
Platt J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Adv Large Margin Classif. 1999;10:61–74.
Molle V, Gulten G, Vilcheze C, Veyron-Churlet R, Zanella-Cleon I, Sacchettini JC, et al. Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis. Mol Microbiol. 2010;78:1591–605.
Delaine T, Bernardes-Génisson V, Quémard A, Constant P, Meunier B, et al. Development of isoniazide NAD truncated adducts embedding a lipophilic fragment as potential bi-substrate InhA inhibitors and antimycobacterial agents. Eur J Med Chem. 2010;45:4554–61.
Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol. 1995;245:43–53.
Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–48.
Acknowledgements
This work was supported in part by a Takeda Science Foundation to JT, and a Grant-in-Aid for Scientific Research (C) (26460145) to SA and (18K05358) to HS from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Taira, J., Umei, T., Inoue, K. et al. Improvement of the novel inhibitor for Mycobacterium enoyl-acyl carrier protein reductase (InhA): a structure–activity relationship study of KES4 assisted by in silico structure-based drug screening. J Antibiot 73, 372–381 (2020). https://doi.org/10.1038/s41429-020-0293-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-020-0293-6