Abstract
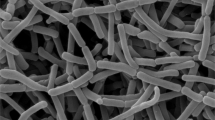
A Gram-staining positive and nonmotile strain designated SYSUP0005T was isolated from tubers of Gastrodia elata Blume. The 16S rRNA gene sequence result showed that strain SYSUP0005T shared highest sequence similarity with the type strain of Amycolatopsis cappadoca (95.7%), Amycolatopsis taiwanensis (95.4%), Amycolatopsis pigmentata (95.4%), Amycolatopsis ruanii (95.1%), and Amycolatopsis helveola (94.8%). Growth occurs at 14–37 °C (optimum temperature, 28 °C), at pH 6–9 (optimum, pH 8) and in the presence of up to 6% (w/v) NaCl. Strain SYSUP0005T had meso-diaminopimelic acid in its peptidoglycan. The whole cell sugars were galactose, ribose, and xylose. The predominant menaquinone was MK-9(H4) and minor menaquinones were MK-9(H2) and MK-9(H8). The polar lipids were diphosphatidylglycerol (DPG); phosphatidylmonomethylethanolamine (PME), phosphatidylethanolamine (PE), phosphatidylinositol (PI), unidentified glycolipid (GL), and unidentified phospholipid (PL). The genomic DNA G + C content was 69.6 mol%. The major fatty acids were iso-C16:0, anteiso-C17:0, C16:0, iso-C14:0, C17:1 ω6c, C17:0, and Summed Feature 3 (C16:1 ω7c/C16:1 ω6c). On the basis of the phenotypic, phylogenetic, chemotaxonomic characters, and genomic comparison, SYSUP0005T represents a novel species of the genus Amycolatopsis, for which the name Amycolatopsis alkalitolerans sp. nov. is proposed. The type strain is SYSUP0005T (=KCTC 49024T = CGMCC4.7463T).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Lechevalier MP, Prauser H, Labeda DP, Ruan JS. Two new genera of nocardioform actinomycetes: Amycolata gen. nov. and Amycolatopsis gen. nov. Int J Syst Bacteriol. 1986;36:29–37.
Thawai C. Amycolatopsis rhizosphaerae sp. nov., isolated from rice rhizosphere soil. Int J Syst Evol Microbiol. 2018;68:1546–51.
Bian J, et al. Amycolatopsis marina sp. nov., an actinomycete isolated from an ocean sediment. Int J Syst Evol Microbiol. 2009;59:477–81.
Işık K, et al. Amycolatopsis cappadoca sp. nov., isolated from soil. Antonie van Leeuwenhoek. 2018;111:1175–82.
Miao Q, et al. Amycolatopsis endophytica sp. nov., a novel endophytic actinomycete isolated from oil-seed plant Jatropha curcas L. Antonie van Leeuwenhoek. 2011;100:333–9.
Huang Y, Pas´ciak M, Liu Z, Xie Q, Gamian A. Amycolatopsis palatopharyngis sp. nov., a potentially pathogenic actinomycete isolated from a human clinical source. Int J Syst Evol Microbiol. 2004;54:359–63.
Wang J, Leiva S, Huang J, Huang Y. Amycolatopsis antarctica sp. nov., isolated from the surface of an Antarctic brown macroalga. Int J Syst Evol Microbiol. 2018;68:2348–56.
Murakami R, et al. A-102395, a new inhibitor of bacterial Translocase I, produced by Amycolatopsis sp. SANK 60206. J Antibiot. 2007;60:690–5.
Dasari VR, Muthyala MK, Nikku MY, Donthireddy SR. Novel Pyridinium compound from marine actinomycete, Amycolatopsis alba var. nov. DVR D4 showing antimicrobial and cytotoxic activities in vitro. Microbiol Res. 2012;167:346–51.
Zhang H, et al. Description of Paracoccus endophyticus sp. nov., isolated from Gastrodia elata Blume. Int J Syst Evol Microbiol. 2019;69:261–5.
Li YQ, et al. Description of Sphingomonas mesophila sp. nov., isolated from Gastrodia elata Blume. Int J Syst Evol Microbiol. 2019. https://doi.org/10.1099/ijsem.0.003263.
Prabhu DM, et al. Sinomonas mesophila sp. nov., isolated from ancient fort soil. J Antibiot. 2015;68:318–21.
Xu P, et al. Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol. 2005;55:1149–53.
Kovacs N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature. 1956;178:703–4.
Gonzalez C, Gutierrez C, Ramirez C. Halobacterium vallismortis sp. nov.: an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol. 1978;24:710–5.
Pridham TG, Gottlieb D. The utilization of carbon compounds by some actinomycetales as an aid for species determination. J Bacteriol. 1948;56:107–14.
Nie GX, et al. Amycolatopsis dongchuanensis sp. nov., a novel actinobacterium isolated from dry-hot valley in Yunnan, south-west China. Int J Syst Evol Microbiol. 2012;62:2650–6.
Li WJ, et al. Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol. 2007;57:1424–8.
Yoon SH, et al. Introducing EzBio-Cloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–7.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–76.
Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:406–16.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–22.
Collins MD, Pirouz T, Goodfellow M, Minnikin DE. Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol. 1977;100:221–30.
Kroppenstedt RM. Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J Liq Chromatogr. 1982;5:2359–67.
Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101.Newark: Microbial ID, Inc; 1990. .
Minnikin DE, Collins MD, Goodfellow M. Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol. 1979;47:87–95.
Collins MD, Jones D. Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4- diaminobutyric acid. J Appl Bacteriol. 1980;48:459–70.
Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–8.
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64.
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91.
Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12.
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31.
Blin K, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;W1:W81–W87.
Tamura T, Ishida Y, Otoguro M, Suzuki K. Amycolatopsis helveola sp. nov. and Amycolatopsis pigmentata sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2010;60:2629–33.
Tseng M, Yang SF, Li WJ, Jiang CL. Amycolatopsis taiwanensis sp. nov., from soil. Int J Syst Evol Microbiol. 2006;56:1811–5.
Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–31.
Acknowledgements
The authors are grateful to Professor Jung-Sook Lee (KCTC, Korea) for kindly providing the reference type strain. This research was supported by Natural Science Foundation of Guangdong Province, PR China (number 2016A030312003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Narsing Rao, M.P., Li, YQ., Zhang, H. et al. Amycolatopsis alkalitolerans sp. nov., isolated from Gastrodia elata Blume. J Antibiot 73, 35–39 (2020). https://doi.org/10.1038/s41429-019-0222-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-019-0222-8
This article is cited by
-
Amycolatopsis acididurans sp. nov., isolated from peat swamp forest soil in Thailand
The Journal of Antibiotics (2021)